- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Optogenetic Mapping of Synaptic Connections in Mouse Brain Slices to Define the Functional Connectome of Identified Neuronal Populations
Published: Vol 7, Iss 1, Jan 5, 2017 DOI: 10.21769/BioProtoc.2090 Views: 11532
Reviewed by: Geoff LauPascal Fossat Ehsan Kheradpezhouh

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Intraepidermal Nerve Fiber Quantification of the Mouse Hind Paw Footpads: A Detailed and Simplified Protocol
Anastasia Yerushkin [...] Amir Dori
Dec 5, 2025 1224 Views

Whole-Mount Immunostaining for the Visual Separation of A- and C-Fibers in the Study of the Sciatic Nerve
Valeriia Ustymenko [...] Nana Voitenko
Dec 5, 2025 1421 Views

Whole-Mount Visualization of Primary Cilia in the Developing Mouse Brain
Oscar Torres Gutierrez [...] Xuecai Ge
Dec 20, 2025 1003 Views
Abstract
Functional connectivity in a neural circuit is determined by the strength, incidence, and neurotransmitter nature of its connections (Chuhma, 2015). Using optogenetics the functional synaptic connections between an identified population of neurons and defined postsynaptic target neurons may be measured systematically in order to determine the functional connectome of that identified population. Here we describe the experimental protocol used to investigate the excitatory functional connectome of ventral midbrain dopamine neurons, mediated by glutamate cotransmission (Mingote et al., 2015). Dopamine neurons are made light sensitive by injecting an adeno-associated virus (AAV) encoding channelrhodopsin (ChR2) into the ventral midbrain of DATIREScre mice. The efficacy and specificity of ChR2 expression in dopamine neurons is verified by immunofluorescence for the dopamine-synthetic enzyme tyrosine hydroxylase. Then, slice patch-clamp recordings are made from neurons in regions recipient to dopamine neuron projections and the incidence and strength of excitatory connections determined. The summary of the incidence and strength of connections in all regions recipient to dopamine neuron projections constitute the functional connectome.
Keywords: ChannelrhodopsinBackground
To establish the function of specific neural circuits it is necessary to determine the anatomical connectome, the mapping of anatomical connections, and its functional connectome, the mapping of the strength, incidence and neurotransmitter nature of connections. The use of viral transsynaptic tracing techniques that are monosynaptically restricted, allows for the description of complex anatomical connections of neural circuits, including the dopamine system (Callaway and Luo, 2015; Faget et al., 2016). The functional connectivity of these circuits has been harder to determine due to the intermingling of axons that make selective electrical stimulation impossible. With the advent of optogenetics it became possible to stimulate genetically defined populations of cells selectively. This allowed for the identification of new connections made by striatal medium spiny neurons (Chuhma et al., 2011), ventral midbrain glutamate neurons (Hnasko et al., 2012; Root et al., 2014) and dopamine/glutamate neurons (Mingote et al., 2015). Moreover, optogenetics used in a systematic and comprehensive manner to map the incidence and strength of connections of specific neuronal populations to defined postsynaptic target regions, determines the functional connectome of defined neuronal populations (Chuhma et al., 2011; Mingote et al., 2015). In this protocol, we describe how to determine the functional connectome of any genetically defined neuronal population. As an example, we focus on the excitatory functional connectome of dopamine neurons, mediated by glutamate cotransmission (Mingote et al., 2015).
Materials and Reagents
- Glass PCR micropipettes with 1 μl marks (Drummond Scientific, catalog number: 5-000-1001-X10 )
- Kimwipe
- Syringe
- Q-tip
- Surgical blades No.11 (Thomas Scientific, catalog number: 3883B59 )
- Needle
- Mice (THE JACKSON LABORATORY, Strain 006660: DATIREScre )
Note: This knock-in mouse expresses cre recombinase under the transcriptional control of the endogenous dopamine transporter (DAT) promoter. To minimize the interference with the DAT promoter function, cre recombinase expression is driven from the 3’ untranslated region via an internal ribosomal entry sequence (IRES). - AAV5-EF1α-DIO-hChR2(H134R)-EYFP (titer: 8x10e-12 virions/ml) (Addgene, catalog number: 20298 ) is used to drive cre-dependent expression of ChR2-EYFP. The adeno-associated virus (AAV) can be obtained under a MTA from Dr. Karl Deisseroth from the vector core at the University of North Carolina; this AAV is serotype 5 and replication-incompetent.
- Paraffin
- 10% bleach solution
- Carprofen (Rimadyl, Zoetis)
- Ketamine HCl (KetaVed, Vedco)
- Xylazine (Akorn, AnaSed®)
- Puralube VET ointment, sterile ocular lubricant (Dechra)
- Lidocaine HCl (40 mg/ml) for local anesthesia (Boehringer Ingelheim)
- Vetbond, n-butyl cyanoacrylate adhesive (3M)
- 70% ethanol solution
- Betadine
- Neosporin
- Saline
Equipment
- Pipette puller (Sutter Instruments, model: P97 )
- NalgeneTM 280 polyurethane tubing (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 8030-0060 )
- NalgeneTM 180 clear plastic PVC tubing (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 8000-0004 )
- Custom valve controller that delivers momentary pulses of 24 V with 5 watts power (modified pulse controller obtained from General Valve Corporation, model: 9-82-902 )
Note: This product has been discontinued. As an alternative, use UltraMicroPumP III (World Precision Instruments, model: UMP3 ) with a TAXIC900 stereotaxic frame, which allows for a precise control of the rate of delivery of the virus. - Air pressure regulator with gauge (General Cryogenics & Specialty Gas Co.)
- Hair clipper (e.g., Wahl Clipper, model: 09916-4301 )
- Heating pad, water-circulating
- Mouse stereotaxic apparatus (Stoeling, catalog number: 51730D )
- Solenoid valve (e.g., Parker Hannifin, part number: 001-0028-900 )
- Scalpel handle (Thomas Scientific, catalog number: 3883H10 )
- Camera (e.g., Microscope, model: 5MP , catalog number: AM7115MZT)
- Video monitor (e.g., Acer)
- Drill (Black & Decker, model: RTX )
Procedure
- Preparation for surgery (time 30-45 min)
- Pull the Drummond glass pipette using the Sutter pipette puller and then by stroking the tip on a Kimwipe break the tip back to a tip diameter between 20-40 μm (see Figure 1).
- Connect the solenoid pulser to the air pressure regulator with polyurethane tubing; connect the wall-mounted ball valve delivering air with polyethylene tubing (Figure 2).
- Backfill the glass pipette with virus solution (2 μl per mouse). Secure the pipette tip to the stereotaxic holder arm, and then connect the pipette using PVC tubing to a syringe attached to a 3-way stopcock valve. Put a drop of a virus solution (at least 2 μl per mouse) on a clean surface covered with paraffin to avoid breaking the pipette tip. Move the stereotaxic holder arm to touch the pipette tip to the drop of virus solution and then pull on the syringe to slowly apply negative pressure and fill the pipette with virus solution. Keep the pipette with the virus solution in a closed box, with wet paper towels in the bottom to maintain humidity, until you are ready to inject. Any surfaces in contact with the virus should be cleaned with 10% bleach solution.
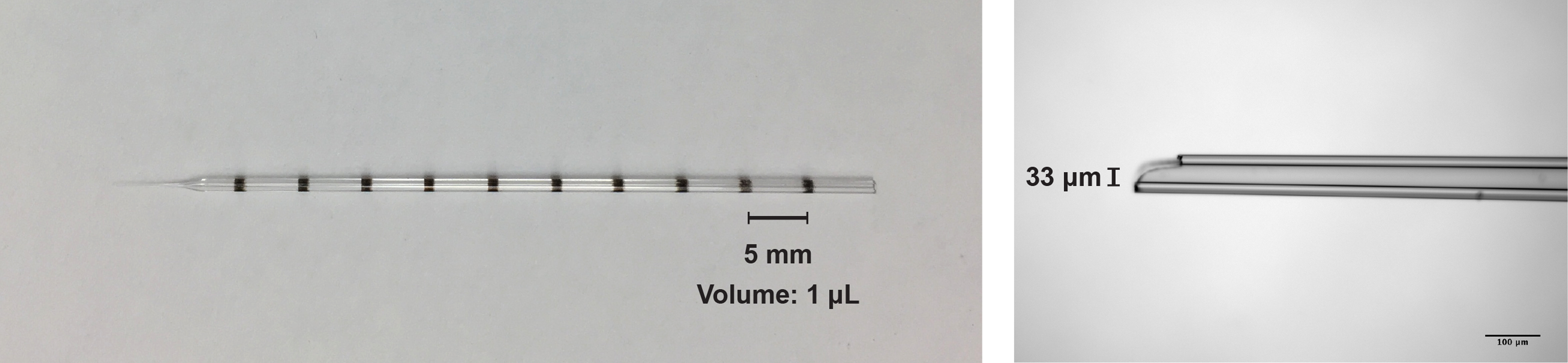
Figure 1. Images of the glass pipette used for the intracranial injection of the virus solution. Left, image of the pipette used for the AAV injection (graduated every 1 μl; maximum 10 μl). Right, photomicrograph of the pipette tip (33 μm diameter).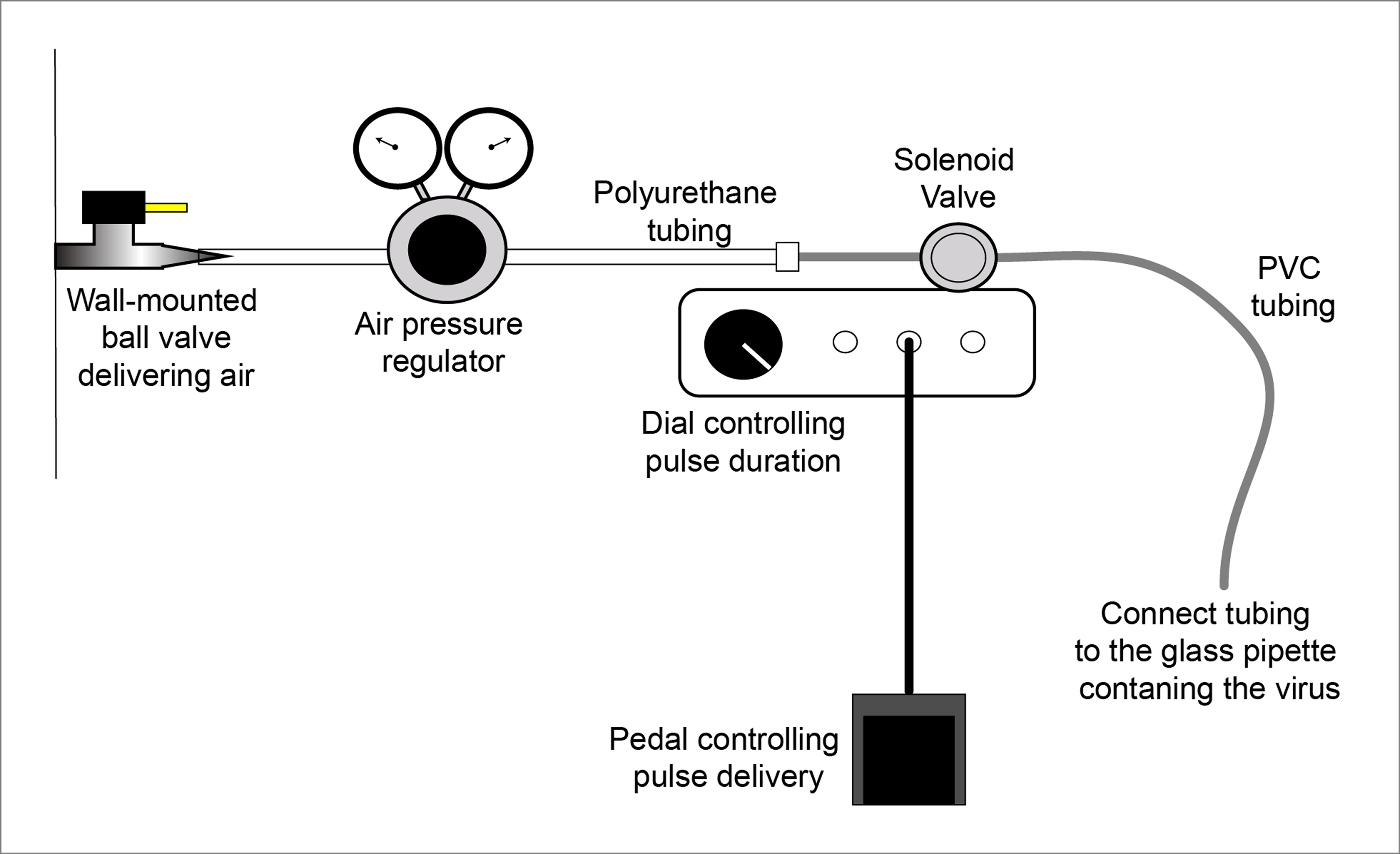
Figure 2. Schematic of the setup using a solenoid valve to deliver time-controlled pulses of compressed air for the administration for the virus through a glass pipette
- Pull the Drummond glass pipette using the Sutter pipette puller and then by stroking the tip on a Kimwipe break the tip back to a tip diameter between 20-40 μm (see Figure 1).
- Surgery and virus injection (time 20-30 min)
- Administer a subcutaneous injection in the neck of caprofen (5 mg/kg) pre-operatively for preemptive analgesia and for its anti-inflammatory actions.
- Anesthetize the mouse with a solution containing ketamine (100 mg/kg) and xylazine (10 mg/kg). If the mouse weighs less than 20 g, administer half the dose. Administer the drugs via intraperitoneal (i.p.) injection at volumes of 10 ml/kg.
- Shave the scalp with the hair clipper.
- Apply Puralube VET ointment to the eyes.
- Place the mouse on a heating pad (37 °C), secure the nose in the mouth bar and nose clamp and stabilize the head using the ear bars.
- Clean the skin above the skull, first with betadine and then with alcohol.
- Apply lidocaine solution using a Q-tip.
- Using a scalpel blade, make an incision and push the skin covering the skull to the side.
- Attach a piece of metal tubing to the stereotaxic probe holder and mark the skull using coordinates targeting the ventral tegmental area (VTA). For mice weighing more than 25 g use the following coordinates: anterior-posterior (AP) - 3.4 mm and lateral (L) -/+ 0.5 mm relative to bregma, and dorsoventral (DV) - 4.5 mm from the dura. Adjust the coordinates to AP - 3.0 mm and DV - 4.1 mm for mice weighing between 12 and 16 g, and to AP - 3.3 mm and DV - 4.3 mm for mice weighing between 17 and 24 g. Do not use the glass pipette to mark the coordinates on the skull, since the tip can easily break.
- Drill holes on both sides and clean the exposed skull with saline.
- Place the glass pipette with the virus solution in the probe holder of the stereotaxic apparatus and connect the pipette to the solenoid valve using PVC tubing (see schematic in Figure 1). Before lowering the pipette into the brain, make sure that the timed solenoid-controlled pulses of compressed air are efficiently ejecting the virus solution. If the virus solution is not coming out, gently clean the pipette tip using a Kimwipe saturated with saline.
- Gently pierce the dura with a needle before lowering the pipette.
- Slowly (approximately 0.2 mm per sec) lower the pipette to the target depth + 0.1 mm. Then raise the pipette 0.1 mm to the target depth (4.3 mm). This creates a pocket for the viral solution and reduces needed pressure when delivering the virus solution. Inject 1 μl of the virus solution per side using timed solenoid-controlled pulses of compressed air.
- Leave the pipette in place for 3 min after the injection to reduce back flux along the injection track, and then withdraw it.
- After both injections, clean the skull with saline, push the skin together with a forceps, and close the scalp incision with Vetbond, allow it to cure (1 min), and then apply Neosporin ointment. Then, remove the animal from the stereotaxic apparatus.
- Administer Carprofen daily for 3 days to alleviate pain.
- Administer a subcutaneous injection in the neck of caprofen (5 mg/kg) pre-operatively for preemptive analgesia and for its anti-inflammatory actions.
Viral transfection delivers multiple copies of ChR2 and induces strong expression. However, ChR2 expression varies among animals so the specificity and efficacy of the transfection needs to be determined and taken into account when determining a functional connectome. Visualization of immunostaining of brains sections for the EYFP tag enables tracing dopaminergic projections, assessing their dopaminergic status and by guiding recordings provides a comprehensive basis for systematic recordings.
Materials and Reagents
- Multi-well plates (Thomas Scientific, catalog number: 1219C16 )
- Petri dishes (Thomas Scientific, catalog number: 1182N84 )
- Coverslips (e.g., Corning, catalog number: 2980-245 )
- Microscope slides coated (e.g., Thomas Scientific, catalog number: 1178T40 )
- Ketamine HCl (KetaVed, Vedco)
- Xylazine (Akorn, AnaSed®)
- Heparin (Swiss Vault Engine, catalog number: 25021-400-66 )
- Phosphate buffer (Sigma-Aldrich, catalog number: P3619 )
- Dulbecco’s phosphate buffer saline (DPBS) (Sigma-Aldrich, catalog number: D5652 )
- Glycine (molecular weight: 75.07) (Sigma-Aldrich, catalog number: 410225 )
- Normal donkey serum (EMD Millipore, catalog number: S30-100mL )
- Triton (Sigma-Aldrich, catalog number: X100 )
- Rabbit polyclonal antibody directed against green fluorescence protein (GFP) (EMD Millipore, catalog number: AB3080 ), which recognizes enhanced yellow fluorescence protein (EYFP)
- Mouse monoclonal antibody against tyrosine hydroxylase (TH) (EMD Millipore, catalog number: MAB318 ), which recognizes a specific marker of dopamine neurons
- Donkey anti-rabbit Alexa Fluor® 488 secondary antibody (Thermo Fisher Scientific, Invitrogen, catalog number: A-21206 )
- Donkey anti-mouse Alexa Fluor® 594 secondary antibody (Thermo Fisher Scientific, Invitrogen, catalog number: A-21203 )
- Prolong Gold mounting medium (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: P36930 )
- Paraformaldehyde (PFA, 16%) (Electron Microscopy Sciences, catalog number: 15710 )
- Glycerol (molecular weight: 92.09) (Sigma-Aldrich, catalog number: G5516 )
- Ethylene glycol (molecular weight: 62.07) (Sigma-Aldrich, catalog number: 324558 )
- Tris HCl (Sigma-Aldrich, catalog number: T2413 )
Equipment
- Brush (small paint brush)
- Vibrating microtome (Leica Biosystems Nussloch, model: VT1200S )
- Laboratory shaker (Benchmark Scientific, model: BlotBoy )
- Slide storage boxes (Thomas Scientific, catalog number: 1202N78 )
- Confocal scanning microscope (Olympus, model: Fluoview FV1000 )
Software
- ImageJ (imagej.nih.gov/ij/download.html) (Abràmoff et al., 2004)
Procedure
- Brain fixation and slicing (1 day)
- After 3-5 weeks post injection, anesthetize mouse with an i.p. injection of a ketamine (100 mg/kg)-xylazine (15 mg/kg) solution.
- Rapidly perfuse intracardially with 1 ml of warm phosphate buffer (30 °C, 0.1 M, pH 7.4) containing 10,000 IU heparin/L, followed by 5 ml of cold phosphate buffer and then by 5 ml of 4% PFA in phosphate buffer.
- Remove the brain and postfix in 4% PFA solution overnight.
- Fill each well of the multi-well plate with 1 ml of cryoprotectant solution.
- Slice the brain on the vibrating microtome at 50 μm to generate coronal sections.
- Collect sections into the multi-well plate.
- Keep brain sections at -20 °C until processing.
- After 3-5 weeks post injection, anesthetize mouse with an i.p. injection of a ketamine (100 mg/kg)-xylazine (15 mg/kg) solution.
- Immunofluorescence staining and imaging (3 days)
- Wash the sections in the multi-well plate with PBS for 5 min in a shaker (12 rpm); repeat this twice.
- Move the sections to a new multi-well plate with glycine (100 mM; 1 ml per well) to quench aldehydes, and put the multi-well plate on the shaker for 30 min.
- Wash sections in PBS for 5 min; repeat this twice.
- Block sections with normal donkey serum (10%) diluted in PBS with 0.1% Triton for 2 h in a shaker. Add 1 ml of this blocking solution per well.
- Dilute the antibodies, anti-GFP (dilution: 1:2,000) and anti-TH (dilution: 1:5,000), in PBS with 2% of normal donkey serum and 0.02% of Triton.
- Move the sections to a new multi-well plate containing both the anti-GFP and anti-TH antibody (1 ml per well), and let the sections incubate on a shaker at 4 °C overnight (minimum 16 h; for better results, leave the antibody incubating over the weekend).
- Wash the sections in PBS for 5 min on the shaker. Repeat this twice.
- Incubate for 2 h with anti-rabbit and anti-mouse secondary antibodies (dilution: 1:200 in PBS-T 0.02%) on the shaker at room temperature.
- Wash the sections in PBS for 5 min; repeat this twice.
- Mount sections on slides in phosphate buffer and let them dry for 5 min.
- When completely dried, put a few drops of the mounting medium on the section and coverslip, being careful not to create bubbles.
- Leave the slides in the dark, overnight at room temperature.
- Apply nail polish around the edges of the coverslip; this is to prevent the coverslip from moving as the mounting medium will not dry.
- Store slides in slide storage boxes at 4 °C. The fluorescence signal typically lasts for 2 years.
- Using a confocal scanning microscope to image the region of interest, in this case the ventral tegmental area and substantia nigra pars compacta (see Figure 1 in Mingote et al., 2015). Use a mouse atlas to make sure that the same regions are imaged for all subjects.
- Take confocal photomicrograph stacks (60x oil objective; optical zoom 1.4x, and z-step increment 0.42 μm; 800 x 800 pixels image frames with a pixel size of 0.189 μm2) through the entire tissue section (40 to 60 images).
- Wash the sections in the multi-well plate with PBS for 5 min in a shaker (12 rpm); repeat this twice.
Data analysis
- Use ImageJ to adjust for contrast and brightness of the confocal stacks linearly.
- Make a z-projected image of the confocal stacks (this image will be used to count) but keep the image with all the confocal stacks also open (this image will be used to identify the different types of cells).
- Identify the number of TH positive (+)/ChR2-EYFP (-), TH (-)/ChR2-EYFP (+) and TH (+)/ChR2-EYFP (+) cells in each image composed of several confocal stacks by scrolling up and down the z-axis and comparing images in the red and green channels.
- Count the number of each type of cell in the z-projected image using the ImageJ cell-counter plug-in.
- Calculate efficacy and specificity percentage values for each stereotaxic position. The efficacy of the virus transfection is calculated as the number of TH (+) and ChR2-EYFP (+) in relation to the total number of TH (+) cells. The specificity of the virus transfection is calculated as the number of TH (+) and ChR2-EYFP (+) in relation to the total number of ChR2 (+) cells (see Figure 3 below, or Figure 2 in Mingote et al., 2015).
- Differences along the anterior-posterior and medial-lateral axes are analyzed using the non-parametric Friedman’s test, with significance set at 0.05.
- Identify the projection areas in other sections of the cells of interest, in this case the ventral midbrain dopamine neurons, which determine the areas to record from.
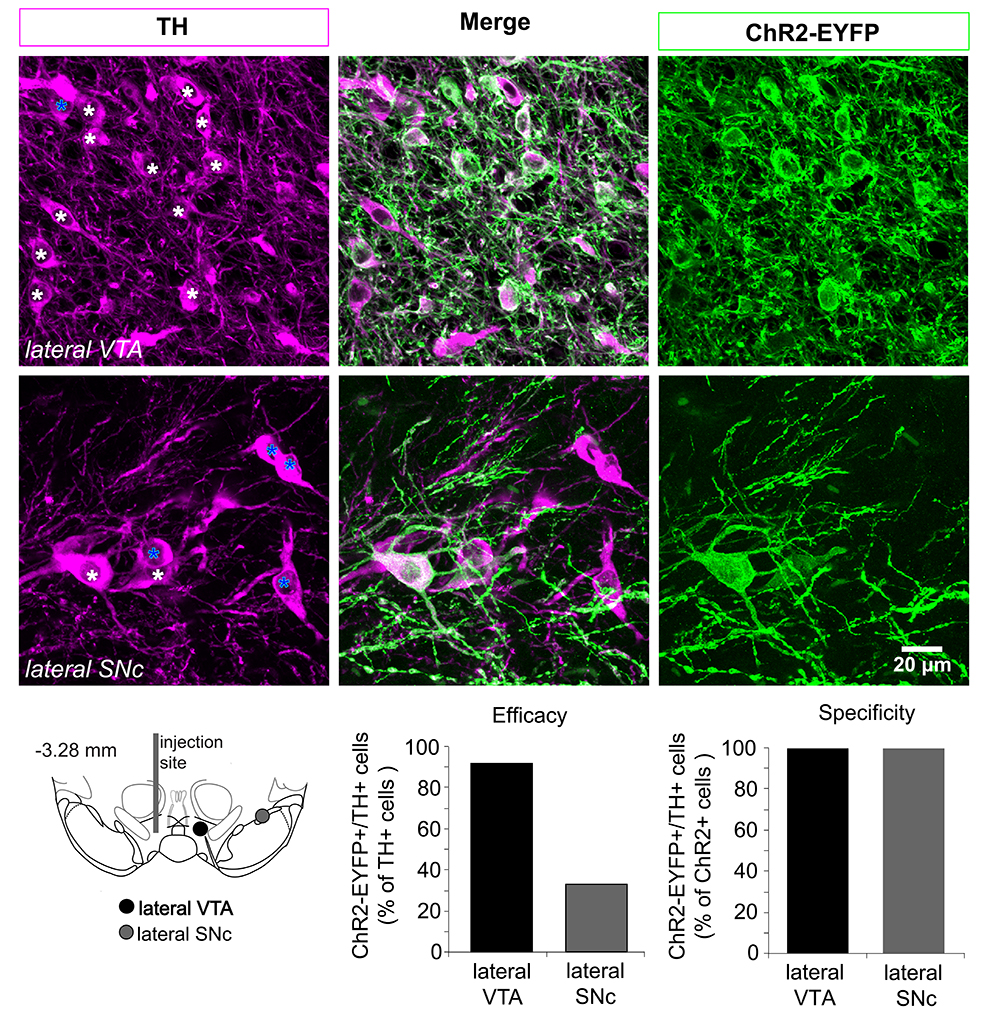
Figure 3. Determining the efficacy and specificity of the ChR2 expression in dopamine neurons. Top panels, confocal z-projected photomicrographs from the lateral VTA and SNc showing cells expressing tyrosine hydroxylase (TH) in magenta, ChR2-EYFP in green, and cells expressing both markers in white. In the panels on the left, white stars indicate TH (+) and ChR2-EYFP (+); blue stars indicate TH (+) and ChR2 (-) cells. Bottom left, Schematic of the ventral midbrain showing the locations where the photomicrographs were taken relative to the AAV injection site. Bottom right, efficacy and specificity graphs based on the cell counts done in the photomicrographs on the top. Efficacy decreases in dopamine neurons located further from the injection site. The specificity using AAV-ChR2-EYFP is very high; the number of TH (-) and ChR2-EYFP- cells is negligible (none are seen here).
Notes
- It has been reported that IgG is present in microglia and this leads to spurious labeling with anti-mouse secondary antibodies; however it has also been shown that such labeling is mouse strain-specific and depends on the immunostaining procedure used (Hazama et al., 2005). Our immunostaining protocol did not produce any non-specific staining of microglia. Alternatively, anti-TH antibody made in chicken or rabbit could be used.
- ChR2 can be expressed in dopamine neurons transgenically, using Ai32 mice (B6;129S-Gt(ROSA)26Sortm32(CAG-COP4*H134R/EYFP)Hze/J;RRID:IMSR_JAX:012569) (Madisen et al., 2012). The transgenic strategy yields reliable expression from animal to animal that cannot be obtained with viral injections (Figure 4). For direct comparison of these two approaches to target dopamine neurons see also Tritsch et al. (2012, supplemental Figure 1). However, the ChR2 specificity in transgenic strategies can be compromised due to ectopic expression. For example, using the serotonin transporter to drive cre expression and ChR2 transgenically (i.e., SERT-cre::Ai32 mice) would not be a good strategy to target Raphe nucleus serotonin neurons selectively since the SERT promotor is transiently active in thalamic cells during development (Narboux-Nême et al., 2008), and those cells would continue to express ChR2 in adulthood.
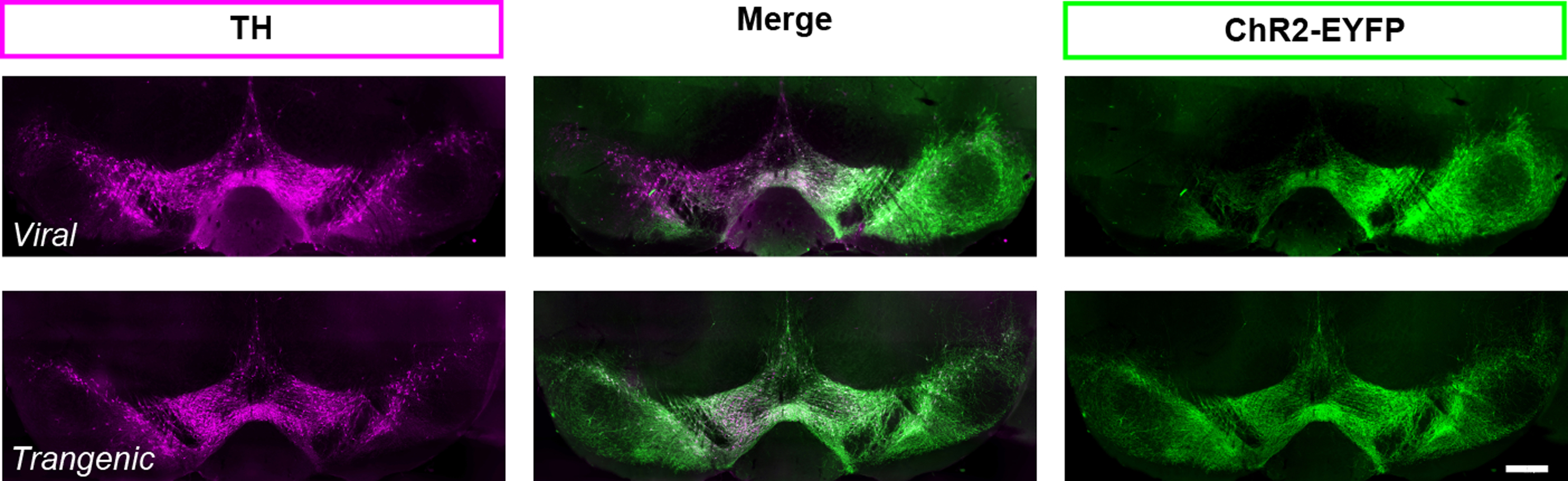
Figure 4. Expression of ChR2-EYFP in TH (+) neurons in the ventral midbrain of DATIREScre mice, induced either by a bilateral injection of an AAV-ChR2-EYFP (top) or breeding with Ai32 mouse (bottom). With both strategies, ChR2 expression is specific to TH (+) cells. However, the viral strategy may not target all the TH (+), as shown on the left side of the photomicrographs in the top panel. When using the viral strategy, ChR2 expression efficacy is highly dependent on the success of the injection and the spread of the virus within the brain region of interest for each injected mouse. For this reason, the efficiency and reliability of expression is much higher with the transgenic strategy (bottom panel). Scale bar represents 400 μm.
Recipes
- 4% paraformaldehyde (PFA) solution (20 ml)
10 ml of 0.2 M phosphate buffer
5 ml of ddH2O
5 ml of 16% PFA - Cryoprotectant solution (100 ml)
30 ml of glycerol
30 ml ethylene glycol
60 ml 0.1 M Tris HCl
Materials and Reagents
- Nalgene 4 mm syringe filter, 0.2 µm pore size, cellulose acetate membrane (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 171-0020 )
- 1 ml Tuberculin slip tip syringe, 1 ml (BD, catalog number: 309659 )
- Nonmetallic syringe needle for filling micropipettes, 28 G, 67 mm (World Precision Instruments, catalog number: MF28G67-5 )
- Pipette tips
- Glass pipette
- Ketamine HCl (KetaVed, Vedco)
- Xylazine (Akorn, AnaSed®)
- Sodium chloride (NaCl) (molecular weight: 58.44) (Sigma-Aldrich, catalog number: 746398 )
- Potassium chloride (KCl) (molecular weight: 74.55) (Sigma-Aldrich, catalog number: 746436 )
- Sodium bicarbonate (NaHCO3) (molecular weight: 84.01) (Sigma-Aldrich, catalog number: S6014 )
- Sodium phosphate monobasic (NaH2PO4) (molecular weight: 119.98) (Sigma-Aldrich, catalog number: S3139 )
- Calcium chloride dehydrate (CaCl2) (molecular weight: 147.01) (Sigma-Aldrich, catalog number: 223506 )
- Magnesium chloride (MgCl2) (molecular weight: 95.21) (Sigma-Aldrich, catalog number: M8266 )
- Glucose (molecular weight: 180.16) (Sigma-Aldrich, catalog number: G6152 )
- Carbogen (95% O2 + 5% CO2)
- SR95531 (gabazine, GABAA antagonist) (molecular weight: 368.23) (Abcam, catalog number: Asc-042 )
- CGP55845 (GABAB antagonist) (molecular weight: 438.71) (Tocris Bioscience, catalog number: 1248 )
- SCH23390 (D1 antagonist) (molecular weight: 324.24) (Tocris Bioscience, catalog number: 0925 )
- (−)-Sulpiride (D2 antagonist) (molecular weight: 341.42) (Tocris Bioscience, catalog number: 0895 )
- Scopolamine hydrobromide (muscarinic antagonist) (molecular weight: 384.27) (Tocris Bioscience, catalog number: 1414 )
- Lidocaine N-ethyl bromide (QX-314, intracellular sodium channel blocker) (molecular weight: 343.30) (Sigma-Aldrich, catalog number: L5783 )
- 6-cyano-7-nitroquinoxaline-2,3-dione disodium salt (CNQX) (molecular weight: 276.12) (Tocris Bioscience, catalog number: 1045 )
- Gluconic acid (molecular weight: 196.16) (Sigma-Aldrich, catalog number: G1951 )
- Cesium hydroxide hydrate (CsOH·H2O) (molecular weight: 149.91) (Sigma-Aldrich, catalog number: C8518 )
- HEPES (molecular weight: 238.30) (Sigma-Aldrich, catalog number: RDD002 )
- EGTA (molecular weight: 380.35) (Sigma-Aldrich, catalog number: E3889 )
- ATP-Na2 (molecular weight: 551.14) (Sigma-Aldrich, catalog number: A2383 )
- GTP-Na2 (molecular weight: 523.18) (Sigma-Aldrich, catalog number: G8877 )
- High-glucose aCSF (see Recipes)
- Standard aCSF (see Recipes)
- Intracellular solution (see Recipes)
Equipment
- Scissors
- Vibrating microtome (Leica Biosystems Nussloch, model: VT1200S )
Note: Different vibratomes are used for histology and for slice recording to avoid detrimental effects of formaldehyde on slice health. - Pipette puller (Sutter Instruments, model: P97 )
- Patch pipettes, standard borosilicate glass capillaries (OD/ID 1.5/0.84 mm) with filament (World Precision Instruments, catalog number: 1B150F-4 )
- CCD camera (Olympus, model: OLY-150 , discontinued), as alternative, scientific CMOS camera with live imaging mode (e.g., Hamamatsu Photonics, model: C11440-42U ) can be used
- High-Power blue LED (470 nm; LED controller current set at 1 A, corresponding to a LED voltage of 10 V) (Thorlabs, model: DC4100 )
- Fluorescence microscope (Olympus, model: BX61WI ) with a 60x water-immersion lens
- Recording chamber (perfusion chamber round) (Siskiyou, model: PC-R )
- Reference electrode (Ag-AgCl electrode, 1 mm diameter, 3 mm long) (World Precision Instruments, catalog number: EP1 )
- Micromanipulator (mechanical, Huxley-style) (Siskiyou, model: MX310R )
- Custom made ‘harp’ to secure slices to recording chamber (U-shaped flattened platinum-iridium wire with single nylon strings attached by cyanoacrylate glue)
- Temperature controller of bath solution (Warner Instruments, model: TC 344B )
- Patch clamp amplifier (Molecular Devices, model: Axopatch 200B )
- Computer for data acquisition with interface (HEKA Elektronik Dr. Schulze, model: InstruTECH ITC-18 ) or (Molecular Devices, model: Axon DigiData 1500 )
Software
- Axograph X (Axograph Scientific) or pClamp 10 (Molecular Devices)
Procedure
- Preparation for slice recording (1 day)
- High-glucose artificial cerebrospinal fluid (aCSF) is used when incubating brain slices; standard aCSF is used for patch-clamp recordings. For both, prepare a 10x stock solutions (see Recipes section).
- Prepare Cesium-gluconate with QX-314 intracellular solution (see Recipes section) and aliquot.
- High-glucose artificial cerebrospinal fluid (aCSF) is used when incubating brain slices; standard aCSF is used for patch-clamp recordings. For both, prepare a 10x stock solutions (see Recipes section).
- Making brain slices and patch-clamp recording (1 day)
- At 3-5 weeks post injection, anesthetize the mouse with ketamine (100 mg/kg) + xylazine (15 mg/kg).
- Open the skull along the sagittal suture with scissors, reflect the skull flaps, and remove the brain into ice-cold high-glucose aCSF (75 mM NaCl, 2.5 mM KCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 0.7 mM CaCl2, 2 mM MgCl2 and 100 mM glucose; see Recipes section), saturated with carbogen.
- Make 300 μm thick slices for each area of interest. To establish the functional connectome of the VTA dopamine neurons, recordings were done in four different coronal sections containing the striatum, the amygdala, the hippocampal formation, or the anterior cortex.
- Preincubate slices in high-glucose aCSF saturated with carbogen for at least 1 h at room temperature for recovery.
- During the incubation time, pull patch pipettes with pipette resistance between 4 and 8 MΩ (tip diameter ~1 µm).
- Transfer one slice to the recording chamber (submerged, 500 μl volume) on the stage of the fluorescence microscope and secure the slice with the harp. The recording chamber should be continuously perfused (1.5 ml/min) with standard aCSF (125 mM NaCl, 2.5 mM KCl, 25 mM NaHCO3, 1.25 mM NaH2PO4, 2 mM CaCl2, 1 mM MgCl2 and 25 mM glucose; see Recipes section) saturated with carbogen, and maintained at 31-33 °C.
- Glutamatergic responses are isolated by blocking dopamine, GABA and acetylcholine actions with a cocktail of antagonists added to the perfusate: 10 μM SR95531, 3 μM CGP55345, 10 μM SCH23390, 10 μM (−)-sulpiride and 1 μM scopolamine.
- Focus on the area of interest using a 10x objective and then use the 60x and a camera to identify the cell to patch. All recordings should be done in areas with visible ChR2-EYFP axons.
- Voltage-clamp recordings are performed with a -75 mV holding potential with the sodium channel blocker (QX-314) in the intracellular solution to block active currents. Cell activity can be monitored in cell attached mode. Once whole-cell mode is achieved, neurons can only be identified based on cell body location, size, and shape, since cesium-based pipette solution including QX-314 blocks intrinsic activity.
- After locating a healthy cell, backfill the patch pipette with the filtered (by 0.2 µm pore syringe filter) cesium-gluconate intracellular solution using the non-metal syringe needle, and mount the pipette in the pipette holder on the micromanipulator. Before immersing the pipette tip in the bath, apply a gentle positive pressure to the glass pipette; this prevents dilution of the intracellular solution due to aspiration of bath solution and clogging of the pipette.
- Advance the pipette to the chosen cell with the micromanipulator. After touching the cell membrane with the tip of the pipette, gently advance to dimple the membrane. Release the positive pressure and confirm that the tip resistance increases, then apply gentle negative pressure to the pipette and apply negative holding potential to achieve a GΩ seal, wait 1-2 min for the seal to maximize. Apply further negative pressure to rupture the membrane at the tip of the pipette for whole cell recording, quickly releasing the negative pressure from the pipette after breaking through.
- To minimize baseline noise for better detection of small amplitude excitatory postsynaptic currents (EPSCs) series resistance is not compensated (see Notes).
- Data acquisition using Axograph X or pClamp commences 5 min after achieving whole-cell mode, to allow for diffusion of intracellular solution into recorded cells.
- Photostimulate with short flashes (5 msec at 0.1 Hz) of a high-power blue LED illumination every 10 sec (0.1 Hz) to evoke synaptic responses. To monitor series resistance, a 5 mV, 5 msec voltage pulse is applied 50 msec before each photostimulation. Record a minimum of 20 responses per cell.
- Glutamatergic responses should be confirmed by perfusion of 40 μM CNQX, allowing at least 5 min after drug application for full effect.
- At 3-5 weeks post injection, anesthetize the mouse with ketamine (100 mg/kg) + xylazine (15 mg/kg).
Data analysis
- For each cell, average 10 consecutive traces. Discard the first trace, since the EPSC amplitude is always significantly larger due to the rested state of the synaptic connection.
- Measure the peak amplitude within the 50 msec post-photostimulation time window.
- The detection threshold for determination of connections should be set to the mean plus 2 standard deviations of the baseline (including spontaneous EPSCs) amplitude in the 100 msec time window preceding the photostimulation. When the peak amplitude of post-photostimulation EPSCs exceeds this threshold, the cell is counted as having a dopamine neuron glutamatergic connection (see Figure 5).
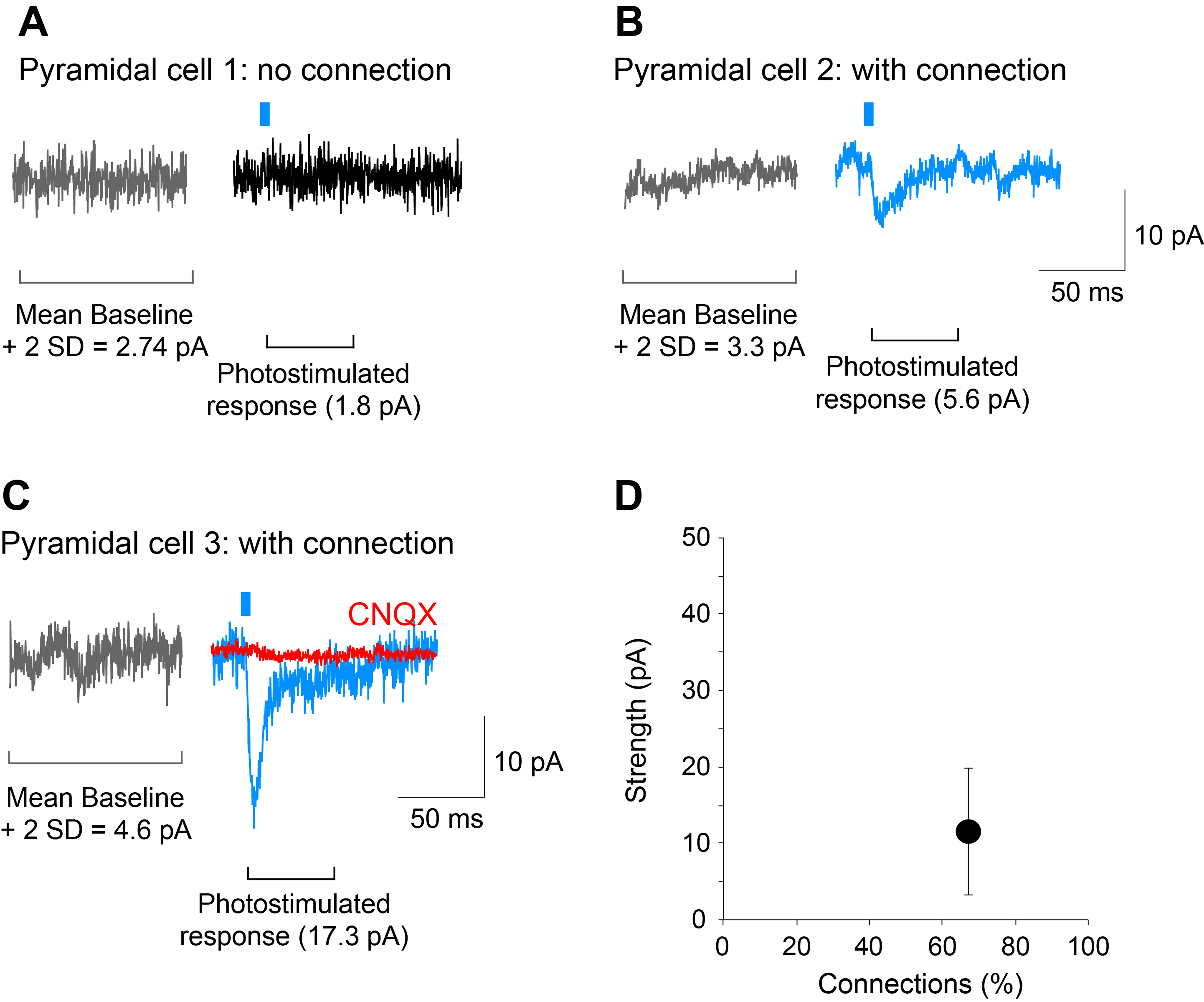
Figure 5. Determining the strength and incidence of the dopamine neuron glutamatergic connections. A-C. Traces showing examples of a cell recorded with no connection (pyramidal cell 1) and two cells with connections of different strengths (pyramidal cell 2 and 3; blue traces). The AMPA/kainate-receptor antagonist CNQX (40 M) completely blocked the photostimulated EPSC (pyramidal cell 3; red trace) showing glutamate mediation. Gray traces were used to calculate the mean amplitude of the baseline plus 2 standard deviations (SD); responses that exceeded this threshold were considered to show connections. D. Graph on the bottom right, is a plot of the data shown in this figure. The strength of the connection is calculated as the mean amplitude of the responses in the two cells with a connection, while the incidence of connections is the percentage of cells that show a connection relative to all cells recorded, in this case, 2 out of 3 cells. Recordings shown are from the entorhinal cortex. Photostimulation is indicated by the blue bar.
Notes
- The bath and pipette solutions generate a ~15 mV liquid junction potential. To correct junction potentials online and to determine the -75 mV holding, set holding command of amplifier to -60 mV (Neher, 1992).
- Since series resistance compensation uses a positive feedback circuit, online compensation increases baseline noise. Generally, if a response is small and slower, series resistance compensation does not affect the amplitude or time course of the response, which we confirmed. However, series resistance compensation should be considered for recordings of larger and faster responses, online or offline (with analysis software).
- To improve the identification of the postsynaptic cell, it is possible to genetically target the recorded cell with GFP or fill the recording cell with a fixable marker (e.g., biocytin, Alexa fluorescent dyes) and then fix the tissue for double labeling of the cell with a known cell marker.
Recipes
- 10x aCSF stock solution without glucose, magnesium or calcium
43.83 g of NaCl
1.86 g of KCl
21.84 g of NaHCO3
1.5 g of NaH2PO4
Add ddH2O to make 1 L solution and store it at 4 °C - High-glucose aCSF (1 L)
On the day of the slice recording experiment make the high-glucose aCSF
Dilute the 10x stock solution 10 times (100 ml of the stock solution in 900 ml of ddH2O)
Add 18.02 g of glucose
Add 2 ml of a 1 M solution of MgCl2 (add 95.21 mg per 1 ml ddH2O)
Add 0.7 ml of a 1 M solution of CaCl2 (add 147 mg per 1 ml ddH2O) - Standard aCSF (1 L)
On the day of the slice recording experiment make the standard aCSF
Dilute the stock solution 10 times (100 ml of the stock solution in 900 ml of ddH2O)
Add 4.5 g of glucose
Add 1 ml of a 1 M solution of MgCl2 (add 95.21 mg per 1 ml ddH2O)
Add 2 ml of a 1 M solution of CaCl2 (add 147 mg per 1 ml ddH2O)
If adding CaCl2 causes precipitation of calcium carbonate, saturate the solution with carbogen to clear the precipitation - Intracellular solution (Cesium-gluconate containing QX-314, 25 ml), Osmolarity 287 mOsm
1 ml of the 3.25 M gluconic acid (molarity of the Sigma-Aldrich solution)
3.25 ml of 1 M CsOH solution (add 130 mg per 1 ml of ddH2O)
50 μl of 1 M MgCl2 solution (add 95.21 mg per 1 ml ddH2O)
2.5 μl of 1 M CaCl2 solution (add 147 mg per 1 ml ddH2O)
2.5 ml of 50 mM QX-314 solution (add 17.2 mg per 1 ml ddH2O)
250 μl of 1 M HEPES solution (add 238.30 mg per 1 ml ddH2O)
9.5 mg of EGTA
Add 17.948 ml of ddH2O
Let the solution sit overnight at 4 °C to stabilize the pH
Adjust the pH to 7.3 with CsOH solution
Add 25.4 mg of ATP-Na2
Add 3.9 mg of GTP-Na2
Filter the solution with a 2 μm filter
Make 1 ml aliquots and store them at -20 °C
Acknowledgments
This protocol was used to obtain the data published in the Journal of Neuroscience (Mingote, S., Chuhma, N., Kusnoor, S. V., Field, B., Deutch, A. Y. and Rayport, S. [2015]). Functional connectome analysis of dopamine neuron glutamatergic connections in forebrain regions. J Neurosci 35[49]: 16259-16271). Procedures involving mice were conducted in accordance with the guidelines of the National Institute of Health Guide for the Care and Use of Laboratory Animals under protocols approved by the Institutional Animal Care and Use Committees of Columbia University and New York State Psychiatric Institute. The work was supported by a NARSAD Young Investigator award (SM), DA017978 and MH087758 (SR).
References
- Abràmoff, M. D., Magalhães, P. J. and Ram, S. J. (2004). Image processing with ImageJ. Biophotonics international 11(7): 36-42.
- Callaway, E. M. and Luo, L. (2015). Monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J Neurosci 35(24): 8979-8985.
- Chuhma, N. (2015). Optogenetic analysis of striatal connections to determine functional connectomes. In: Hiromu, Y. (Ed.). Optogenetics. Springer pp: 265-277.
- Chuhma, N., Tanaka, K. F., Hen, R. and Rayport, S. (2011). Functional connectome of the striatal medium spiny neuron. J Neurosci 31(4): 1183-1192.
- Faget, L., Osakada, F., Duan, J., Ressler, R., Johnson, A. B., Proudfoot, J. A., Yoo, J. H., Callaway, E. M. and Hnasko, T. S. (2016). Afferent inputs to neurotransmitter-defined cell types in the ventral tegmental area. Cell Rep 15(12): 2796-2808.
- Hazama, G. I., Yasuhara, O., Morita, H., Aimi, Y., Tooyama, I. and Kimura, H. (2005). Mouse brain IgG-like immunoreactivity: strain-specific occurrence in microglia and biochemical identification of IgG. J Comp Neurol 492(2): 234-249.
- Hnasko, T. S., Hjelmstad, G. O., Fields, H. L. and Edwards, R. H. (2012). Ventral tegmental area glutamate neurons: electrophysiological properties and projections. J Neurosci 32(43): 15076-15085.
- Madisen, L., Mao, T., Koch, H., Zhuo, J. M., Berenyi, A., Fujisawa, S., Hsu, Y. W., Garcia, A. J., 3rd, Gu, X., Zanella, S., Kidney, J., Gu, H., Mao, Y., Hooks, B. M., Boyden, E. S., Buzsaki, G., Ramirez, J. M., Jones, A. R., Svoboda, K., Han, X., Turner, E. E. and Zeng, H. (2012). A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci 15(5): 793-802.
- Mingote, S., Chuhma, N., Kusnoor, S. V., Field, B., Deutch, A. Y. and Rayport, S. (2015). Functional connectome analysis of dopamine neuron glutamatergic connections in forebrain regions. J Neurosci 35(49): 16259-16271.
- Narboux-Nême, N., Pavone, L. M., Avallone, L., Zhuang, X. and Gaspar, P. (2008). Serotonin transporter transgenic (SERTcre) mouse line reveals developmental targets of serotonin specific reuptake inhibitors (SSRIs). Neuropharmacology 55(6): 994-1005.
- Neher, E. (1992). Correction for liquid junction potentials in patch clamp experiments. Methods Enzymol 207: 123-131.
- Root, D. H., Mejias-Aponte, C. A., Zhang, S., Wang, H. L., Hoffman, A. F., Lupica, C. R. and Morales, M. (2014). Single rodent mesohabenular axons release glutamate and GABA. Nat Neurosci 17(11): 1543-1551.
- Tritsch, N. X., Ding, J. B. and Sabatini, B. L. (2012). Dopaminergic neurons inhibit striatal output through non-canonical release of GABA. Nature 490(7419): 262-266.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Mingote, S., Chuhma, N. and Rayport, S. (2017). Optogenetic Mapping of Synaptic Connections in Mouse Brain Slices to Define the Functional Connectome of Identified Neuronal Populations. Bio-protocol 7(1): e2090. DOI: 10.21769/BioProtoc.2090.
-
Mingote, S., Chuhma, N., Kusnoor, S. V., Field, B., Deutch, A. Y. and Rayport, S. (2015). Functional connectome analysis of dopamine neuron glutamatergic connections in forebrain regions. J Neurosci 35(49): 16259-16271.
Category
Neuroscience > Cellular mechanisms > Synaptic physiology
Neuroscience > Neuroanatomy and circuitry > Immunofluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








