- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Ubiquitin Dimer Formation Assay
Published: Vol 7, Iss 1, Jan 5, 2017 DOI: 10.21769/BioProtoc.2082 Views: 8645
Reviewed by: Yanjie LiQiangjun ZhouAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1465 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3196 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1812 Views
Abstract
The process of protein ubiquitination typically consists of three sequential steps to add an ubiquitin (Ub) or Ub chain to a substrate protein, requiring three different enzymes, ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2), and ubiquitin protein ligase (E3). Most E2s possess the classical E2 activity in forming E2-Ub complex through a thioester linkage, in presence of an E1 and Ub. Additionally, some E2s have the ability of catalyzing the formation of free Ub dimer. Such activity indicates an important role of these E2s in ubiquitination pathway. Thus, we developed an in vitro Ub dimer formation assay to determine the activity of certain E2s. Moreover, by using Ub mutants, in which different lysine residues are mutated, the specific linkage of dimer can also be determined.
Keywords: UbiquitinationBackground
The existing protocols for E2 conjugation initiation assay (without adding E3 and substrate) aim to detect the thioester linkage (E2-S-Ub). Our method focuses on the E2 activity of catalyzing free Ub dimer formation (Ub-Ub). It provides a convenient way to detect an important biochemical feature of E2 in different species. Further, the specific linkage of dimer can be determined by using different Ub mutants.
Materials and Reagents
- 1.5 ml polypropylene tubes
- Polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories, catalog number: 162-0177 )
- AmershamHyperfilmTM ECL (GE Healthcare, catalog number: 28906836 )
- Purified human recombinant E1 (BOSTONBIOCHEM, catalog number: K-995 )
- Qiagen Ni-NTA Spin Kit (QIAGEN, catalog number: 31314 )
- Purified human recombinant Ub (BOSTONBIOCHEM, catalog number: K-995 )
- Purified human recombinant Ub with the lysine 11 (K11) residue mutated (Ub-K11R) (BOSTONBIOCHEM, catalog number: UM-K11R )
- Purified human recombinant Ub with the lysine 48 (K48) residue mutated (Ub-K48R) (BOSTONBIOCHEM, catalog number: UM-K48R )
- Purified human recombinant Ub with the lysine 63 (K63) residue mutated (Ub-K63R) (BOSTONBIOCHEM, catalog number: UM-K63R )
- 10x reaction buffer (BOSTONBIOCHEM, catalog number: K-995 )
- Mg-ATP solution (BOSTONBIOCHEM, catalog number: K-995 )
- 4x non-reducing loading buffer (BOSTONBIOCHEM, catalog number: K-995 )
- SDS-PAGE gel
- Skimmed milk powder
- Anti-ubiquitin antibody (Cell Signaling Technology, catalog number: 3936 )
- Goat anti-mouse antibody conjugated to horseradish peroxidase (HRP) (Bio-Rad Laboratories, catalog number: 170-6516 )
- Amersham ECL prime Western blotting detection reagent (GE Healthcare, catalog number: RPN2232 )
- NaCl
- KCl
- Na2HPO4
- KH2PO4
- Tween-20 (Sigma-Aldrich, catalog number: P1379 )
- Tris base
- Glycine
- Methanol
- Dialysis buffer (see Recipes)
- 1x PBS (see Recipes)
- 1x PBST (see Recipes)
- Transfer buffer (see Recipes)
Equipment
- Incubator (VWR, model number: 1545 ) or water bath
- Protein electrophoresis apparatus (Bio-Rad Laboratories, model: Mini PROTEAN® 3 Cell )
- Western blotting apparatus (Bio-Rad Laboratories, model: Mini Trans-Blot® Cell )
- X-Ray film processor (PROTEC, model: OPTIMAX )
Procedure
- Purify 6x His-tagged Arabidopsis E2 UBC22 (His-UBC22) using QIAGEN Ni-NTA Spin Kit and following the manufacturer’s instructions. Purified protein is then dialyzed in a dialysis buffer for overnight.
- Prepare the reaction samples in 1.5 ml polypropylene tubes. Different components are added as listed below. To test specific linkage of dimer, wild-type Ub can be substituted by different Ub mutants, each of which has one particular lysine residue mutated (Ub-K11R, Ub-K48R or Ub-K63R).
Component Amount to add or final concentration
10x buffer 2.0 μl
E1 0.2 μM
E2 5.0 μM
Ub 62.5 μM
Mg-ATP solution 1.0 mM
ddH2O make up to 20 μl - Incubate the reaction samples at 30 °C either in a water bath or an incubator for 4 h.
- Add 7 μl 4x SDS non-reducing loading buffer and 1 μl ddH2O into each reaction sample.
- Treat the samples at 95 °C for 5 min.
- Bio-Rad Mini PROTEAN® 3 Cell gel apparatus is used for protein electrophoresis. Load half amount of samples on a 15% SDS-PAGE resolving gel with 4% stacking gel and run the gel at 120 V for 130 min to separate the proteins.
- Transfer the proteins to a piece of PVDF membrane at 120 V for 120 min using Bio-Rad Mini Trans-Blot® Cell. The transfer apparatus is placed into a bucket with ice to reduce heat production during the transfer.
- Incubate the membrane in 10 ml of 1x PBS containing 5% skimmed milk powder for 0.5 h at room temperature for the blocking treatment.
- Incubate the membrane with primary antibody (anti-ubiquitin antibody, antibody dilution ratio: 1:40,000) in 10 ml 1x PBST containing 3% skimmed milk powder overnight at 4 °C.
- Wash the membrane with 1x PBST for three times at room temperature (20 ml, 10 min each).
- Incubate the membrane with secondary antibody (goat anti-mouse antibody conjugated to HRP, antibody dilution ratio: 1:7,000) in 1x PBST containing 3% skimmed milk powder for 1 h at room temperature.
- Wash the membrane with 1x PBST for three times at room temperature (20 ml, 10 min each).
- Detect signal with the ECL prime Western blotting detection reagent according to the manufacturer’s instructions (Figure 1). Use 1-10 min as the initial range of film exposure time, depending on signal strength.
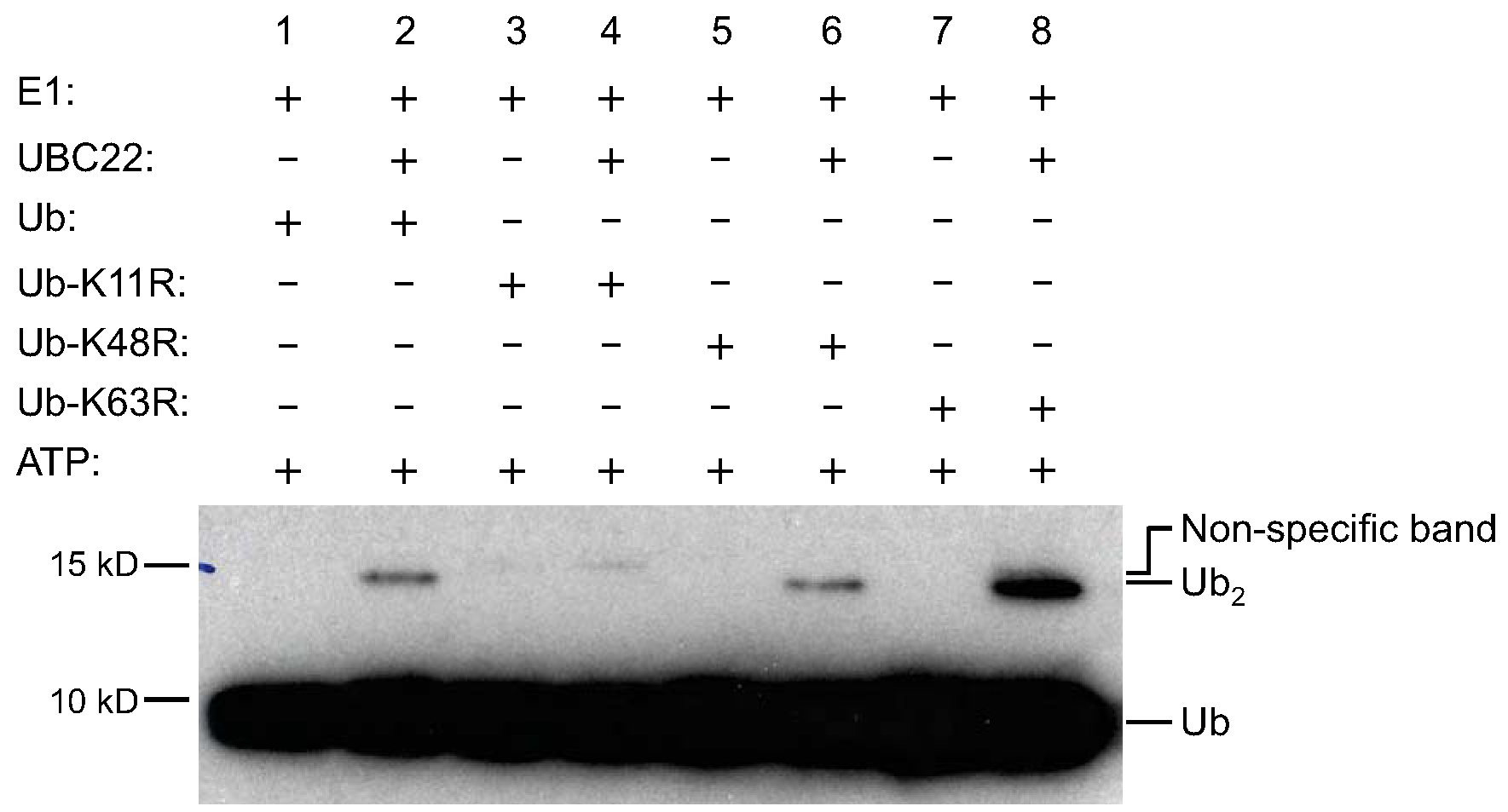
Figure 1. Ub dimer formation assay by His-UBC22. To determine the activity of Arabidopsis UBC22, various components, as indicated on the top of the figure, were added to the reaction tubes with (lanes 2, 4, 6 and 8) or without His-UBC22 (lanes 1, 3, 5 and 7). The reaction tubes were incubated at 30 °C for 4 h. The samples were then subjected to SDS-PAGE (with 4% stacking and 15% resolving gel).Free Ub and Ub dimers were detected by Western blotting using an anti-Ub antibody. Free Ub (Ub) and Ub dimers (Ub2) are indicated on the right side of the figure. For the K11R mutant, no new Ub dimer formation was observed when His-UBC22 protein was added. Due to slight impurity, there was one weak band in the recombinant K11R protein which is slightly higher than the Ub dimer synthesized from the wild-type Ub and other Ub mutants. Three independent experiments were performed and had similar results (Figure from Wang et al., 2016; Figure 6 in the manuscript: http://jxb.oxfordjournals.org/content/early/2016/04/10/jxb.erw142.full).
Data analysis
Purified Arabidopsis recombinant E2 UBC22 fused with 6x His tag (His-UBC22) was tested in the in vitro Ub dimer formation assay. As shown in Figure 1, Ub dimers could be detected when His-UBC22 was added into the reaction (lane 2 compared to the lane 1), indicating the biochemical activity of UBC22 catalyzing free dimer formation. In addition, similar dimer formation was observed when the Ub-K48R mutant or Ub-K63R mutant was used, which lacks K48 residue or K63 residue (lane 6 compared to lane 5, or lane 8 compared to lane 7). Interestingly, when the Ub-K11R which lacks K11 residue was used, little dimer was produced (lane 4 compared to lane 3). These results indicate that UBC22 could catalyze Ub dimer formation in vitro specifically through K11 residue of Ub. Three independent experiments were performed and produced similar results.
Notes
- This assay aims to investigate a specific activity of the E2 enzyme. The purity of the recombinant protein or any conditions affecting a protein’s activity could affect the result. A commercial E2 protein, such as Human E2 Ube2S, may be good for a positive control.
- Poly-Ub chains might be formed and detected in the assay depending on the activity of an E2 tested.
Recipes
- Dialysis buffer
10 mM Tris-HCl (pH 7.5)
50 mM NaCl - 1x PBS
137 mM NaCl
2.7 mM KCl
10 mM Na2HPO4
1.8 mM KH2PO4
Adjust to pH 7.4 - 1x PBST
0.1% Tween-20 in 1x PBS - Transfer buffer
25 mM Tris base
192 mM glycine
20% (v/v) methanol
Acknowledgments
We gratefully acknowledge the financial support from the Natural Sciences and Engineering Research Council of Canada (NSERC) (Discovery grant) to HW.
References
- Wang, S., Cao, L. and Wang, H. (2016). Arabidopsis ubiquitin-conjugating enzyme UBC22 is required for female gametophyte development and likely involved in Lys11-linked ubiquitination. J Exp Bot 67(11): 3277-3288.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, S., Cao, L. and Wang, H. (2017). In vitro Ubiquitin Dimer Formation Assay. Bio-protocol 7(1): e2082. DOI: 10.21769/BioProtoc.2082.
Category
Plant Science > Plant biochemistry > Protein > Modification
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









