- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Gene Expression Analysis of Sorted Cells by RNA-seq in Drosophila Intestine
Published: Vol 6, Iss 24, Dec 20, 2016 DOI: 10.21769/BioProtoc.2079 Views: 16272
Reviewed by: Jihyun KimLeonardo G. GuilgurModesto Redrejo-Rodriguez

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparation of Drosophila Larval Blood Cells for Single-cell RNA Sequencing
Sudhir Gopal Tattikota and Norbert Perrimon
Aug 20, 2021 3778 Views

Preparation of a Single-cell Suspension from Drosophila Wing Imaginal Discs
Shu Yang [...] Xiaoyan Zheng
Aug 20, 2022 2631 Views

Optimized Method for High-Quality Isolation of Single-Nuclei From Mosquito Fat Body for RNA Sequencing
Stephanie Serafim de Carvalho [...] Carolina Barillas-Mury
Jan 5, 2026 249 Views
Abstract
RNA sequencing (RNA-seq) has become a popular method for profiling gene expression. Among many applications, one common purpose is to identify differentially expressed genes and pathways in different biological or pathological conditions. This protocol provides detailed procedure for RNA-seq analysis of ~250,000 sorted Drosophila intestinal cells (Chen et al., 2016), in which RNA amplification is not required.
Keywords: RNA-seqBackground
Transcriptome analysis by RNA-seq has become a popular method for the identification of differentially expressed genes and pathways under different biological or pathological conditions. For samples that yield low mRNA levels, RNA or cDNA amplification was commonly performed before deep-sequencing (Dutta et al., 2015). However, this procedure could potentially omit important candidates that are expressed in low abundance. Here we provide a detailed procedure for RNA-seq analysis of sorted Drosophila gut cells in which RNA amplification is not required.
Materials and Reagents
- Isolation of intestinal progenitor cells by FACS
- Microcentrifuge tube (Corning, Axygen®, catalog number: MCT-150-C )
- 40 μm filters
- 70 μm filters (Corning, Falcon®, catalog number: 352350 )
- The fly strains carrying Esg-GFP fluorescent marker
- Elastase (Sigma-Aldrich, catalog number: E0258 )
- RNAiso Plus (Takara Bio, catalog number: 9108/9109 )
- NaCl
- KCI
- Na2HPO4
- KH2PO4
- Diethyl pyrocarbonate (DEPC) (Sigma-Aldrich, catalog number: D5758 )
- 1x DEPC-PBS (see Recipe)
- Elastase solution (see Recipe)
- Microcentrifuge tube (Corning, Axygen®, catalog number: MCT-150-C )
- RNA isolation
- Directzol RNA MiniPrep Kit (ZYMO RESEARCH, catalog number: R2050 )
- 95-100% ethanol
- Directzol RNA MiniPrep Kit (ZYMO RESEARCH, catalog number: R2050 )
- Library construction
- Agilent RNA 6000 Pico Kit (Agilent Technologies, catalog number: 5067-1513 )
- Dynabeads® mRNA DIRECTTM Purification Kit (Thermo Fisher Scientific, AmbionTM, catalog number: 61011 )
- Hexadeoxyribonucleotide mixture, pd(N)6 (Takara Bio, catalog number: 3801 )
- dNTP mixture (Takara Bio, catalog number: 4019 )
- DTT (provided by SuperScript® II reverse transcriptase)
- Recombinant RNasin® ribonuclease inhibitor (Promega, catalog number: N2511 )
- SuperScript® II reverse transcriptase (Thermo Fisher Scientific, InvitrogenTM, catalog number: 18064014 )
- Second-strand buffer (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10812014 )
- DNA polymerase I (Takara Bio, catalog number: 2130A )
- RNase H (New England Biolabs, catalog number: M0297S )
- Agencourt AMPure XP Kit (5 ml) (Beckman Coulter, catalog number: A63880 )
- NEBNext® DNA library prep master mix set for Illumina® (New England Biolabs, catalog number: E6040L )
- NEBNext® multiplex oligos for Illumina® (New England Biolabs, catalog number: E7335S / E7500S )
- Agilent High Sensitivity DNA Kit (Agilent Technologies, catalog number: 5067-4626 )
- Qubit® dsDNA HS Assay Kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q32851 )
- Illumina Library Quantification Kit (Kapa Biosystems, catalog number: KK4824 )
- Agilent RNA 6000 Pico Kit (Agilent Technologies, catalog number: 5067-1513 )
Equipment
- Isolation of intestinal progenitor cells by FACS
- Dissecting microscope with zoom and dual goosenecks to supply oblique illumination; CO2 equipped fly sorting station (Leica, model: MZ16 ; custom fabrication)
- Forceps, Dumont #5 (Fine Science Tools, catalog number: 11252-30 )
- Dissecting dish (Thermo Fisher Scientific, Fisher Scientific, catalog number: 21-379 )
- FACS Aria II sorter (BD)
- RNA isolation
- Vortex
- Vortex
- Library construction
- DynaMagTM-2 magnet (Thermo Fisher Scientific, catalog number: 12321D )
- Agencourt AMPure XP 5 ml Kit (Beckman Coulter, catalog number: A63880 )
- PCR thermal cycler
- Qubit® 3.0 Fluorometer (Thermo Fisher Scientific, InvitrogenTM, model: Qubit® 3.0 Fluorometer)
- Agilent 2100 Bioanalyzer (Agilent Technologies, model: 2100 Bioanalyzer)
- Applied BiosystemsTM 7500 Fast & 7500 real-time PCR system (Thermo Fisher Scientific, model: 7500 Fast & 7500 real-time PCR system)
- Hiseq-2500 sequencing system (Illumina, model: Hiseq-2500)
- DynaMagTM-2 magnet (Thermo Fisher Scientific, catalog number: 12321D )
Software
- Mapping and analysis of Illumina reads for transcriptome
- Bowtie (http://bowtie-bio.sourceforge.net/index.shtml, Langmead et al., 2009)
- TopHat (http://ccb.jhu.edu/software/tophat/index.shtml, Trapnell et al., 2009)
- Cufflinks (http://cole-trapnell-lab.github.io/cufflinks/, Roberts et al., 2011)
- CASAVA (http://support.illumina.com.cn/sequencing/sequencing_software/casava.html, Illumina)
- Bowtie (http://bowtie-bio.sourceforge.net/index.shtml, Langmead et al., 2009)
Procedure
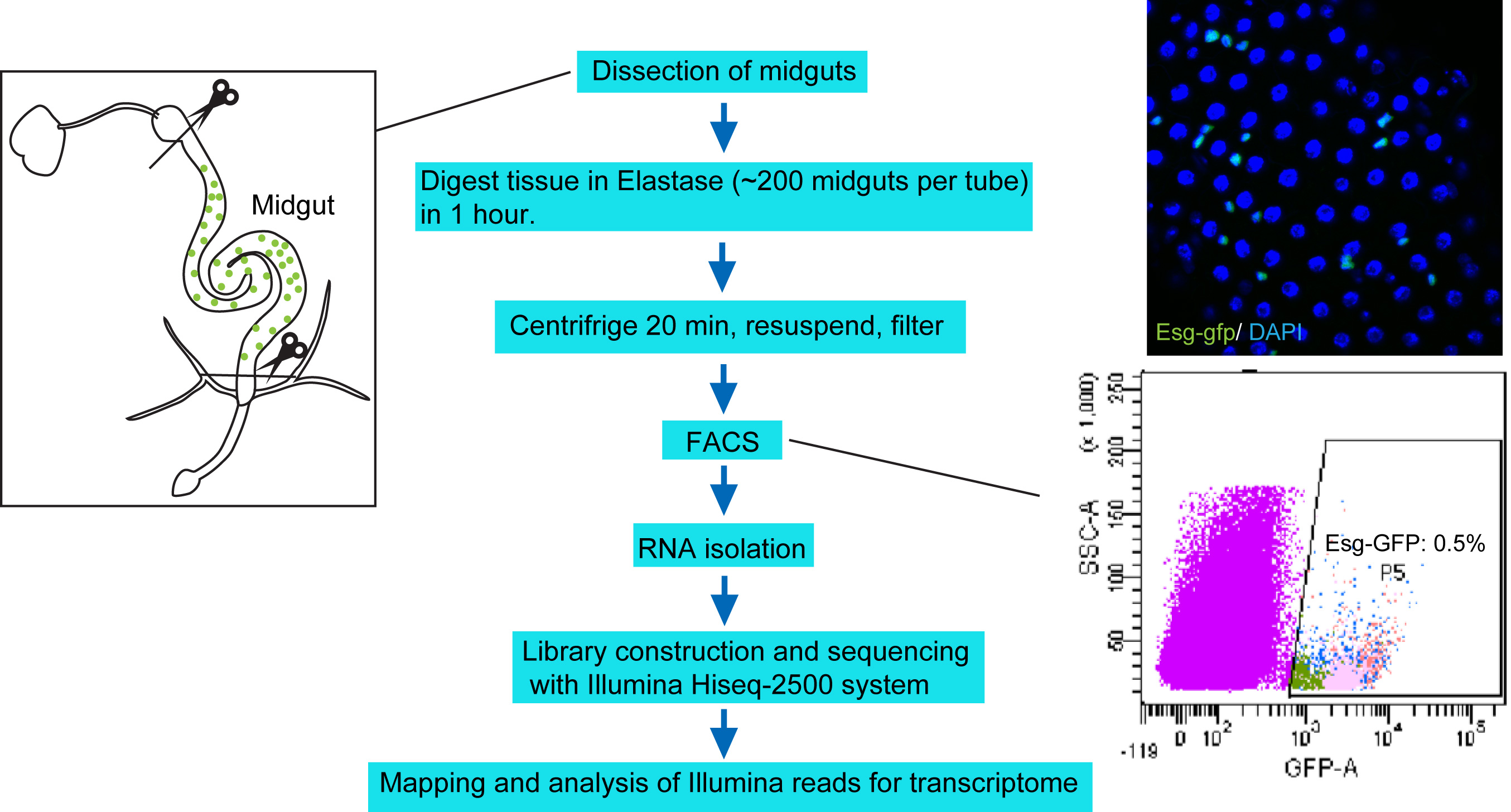
Figure 1. Protocol overview. Under RNase-free environment, the midguts with foregut and hindgut portion removed (left panel) are digested with Elastase for 1 h. The dissociated tissues are centrifuged, resuspended in DEPC-PBS, filtered, and then sorted through FACS. About 250,000 sorted cells are collected to harvest total RNA, which is then used for library construction and sequencing with Illumina Hiseq-2500 system. The upper right panel shows a confocal image of midgut epithelium with Esg-GFP expression. The bottom right panel highlights the Esg-GFP+ cell population measured by FACS.
- Isolation of intestinal progenitor cells by FACS (Figure 1)
- The fly strains carrying Esg-GFP fluorescent markers are used to sort intestinal progenitor cell population (including intestinal stem cells and enteroblasts), midguts from w1118 strain are used to set fluorescence gate. To achieve 250K Esg-GFP+ cells, a minimum of 1,000 midguts is required.
Note: It is important to be consistent with age, gender and culture conditions among different biological samples. Here we take expression analysis of Sox21a mutant flies for example. Sox21a mutants carrying Esg-GFP markers served as the mutant sample, with Esg-GFP wild-type flies as WT Ctrl. Based on the observation that overproliferation phenotype is displayed after 10 days in 90% sox21a mutant intestines, we analyzed flies at 10 days old for both the mutants and WT control. For both groups, we chose female flies and cultured them with identical food (standard fly food plus yeast paste). - The following steps are performed under RNase-free conditions. Forceps and the dissecting pads are pre-washed with DEPC-PBS solution. The prepared females are ice-anesthetized for dissection. Intestines are dissected and the foregut and hindgut parts are removed by forceps (Figure 2). The midguts are then immediately put in DEPC-PBS (see Recipes) solution on ice.
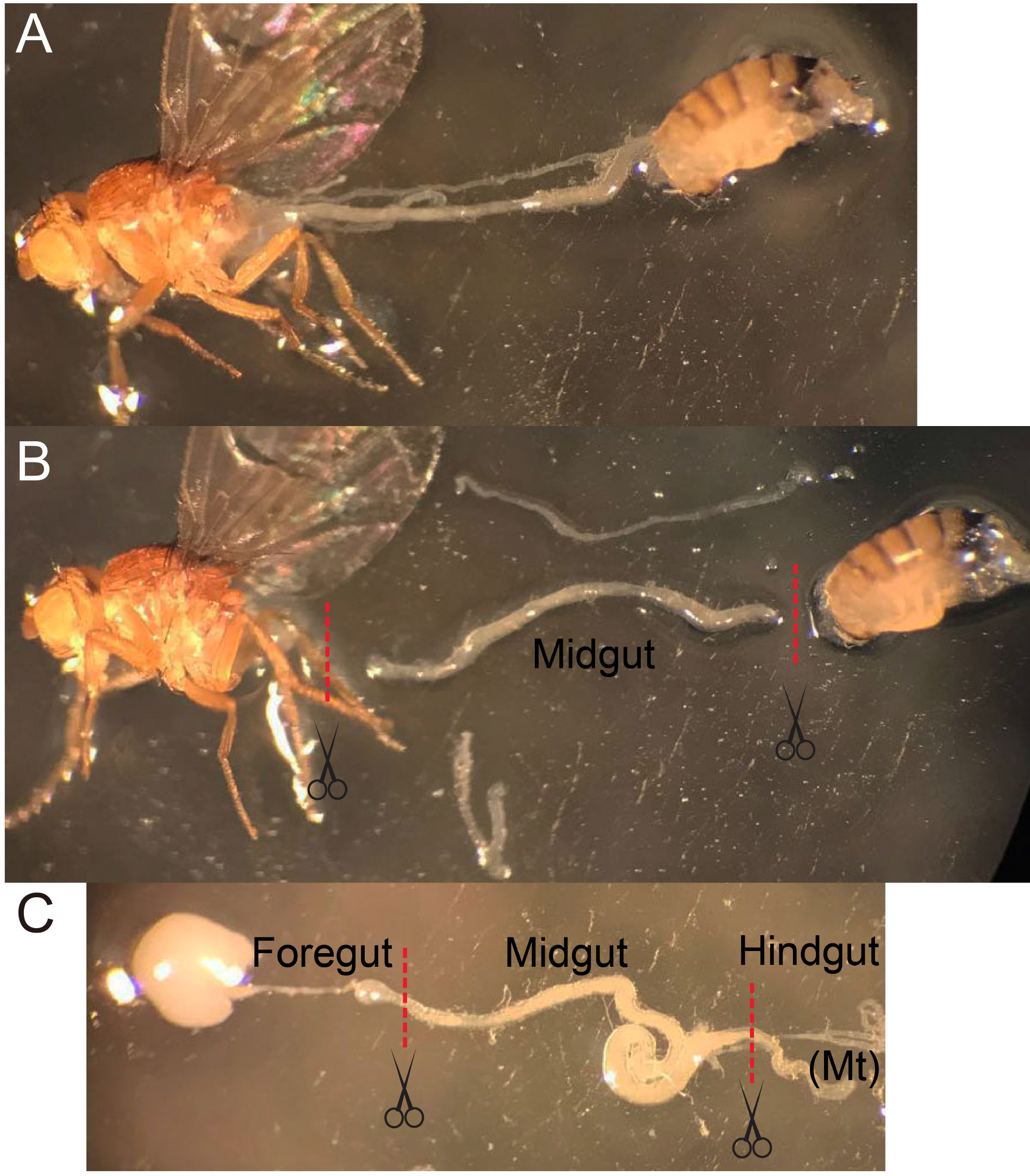
Figure 2. The midgut dissection procedure. A. Use forceps to gently hold a fly and tear the abdomen at the boundary of thorax/abdomen, pull the abdomen away from the anterior part without touching the gut. B and C. Before the gut is fully stretched, cut the gut at the boundaries between foregut and hindgut. Remove the appendix if Malpighian tubule (Mt) or ovarium is attached to the midgut. - Incubate 100-200 guts with 1 mg/ml Elastase (see Recipes) in about 1 ml DEPC-PBS per microcentrifuge tube for 1 h at 25 °C until the midguts are largely dissociated. Softly mix the sample every 15 min by pipetting and inverting several times.
- Dissociated samples are pelleted at 400 x g for 20 min, and resuspended in a microcentrifuge tube with 0.5 ml DEPC-PBS. The suspension was filtered with 40 or 70-μm filters (Corning), by touching pipette tips on the top of the filter so that the cells can go through filters. Wash the tubes and the filters with 0.5 ml DEPC-PBS, and also collect the filtered suspension in the filtered cells. The filtered cells are then sorted using a FACS Aria II sorter (BD Biosciences) and collected in 0.5 ml DEPC-PBS.
Note: 40-μm filters are recommended for pure isolation of small diploid cells, such as intestinal progenitor cells or enteroendocrine cells. It’s recommended to double check the sorted cells based on cell morphology and expression of the fluorescent marker. - The sorted cells are pelleted at 400 x g for 20 min and preserved in 0.2-0.8 ml RNAiso Plus (Takara Bio) reagents, which can be stored at -80 °C within 6 months until RNA isolation. For each of the biological replicates, at least 200,000 sorted cells are collected to harvest total RNA. For each genotype group, at least 3 biological replicates are prepared.
- The fly strains carrying Esg-GFP fluorescent markers are used to sort intestinal progenitor cell population (including intestinal stem cells and enteroblasts), midguts from w1118 strain are used to set fluorescence gate. To achieve 250K Esg-GFP+ cells, a minimum of 1,000 midguts is required.
- RNA isolation
- Add equal volume of > 95% ethanol directly to the homogenate cell sample. Mix thoroughly by vortexing.
- Follow the Directzol RNA MiniPrep Kit to harvest total RNA of each sample. DNase I treatment is unnecessary, as polyA RNAs will be selectively purified during cDNA library construction. The RNA sample eluted in DNase/RNase-free water should be used immediately for sequencing library construction or stored at -80 °C for up to 3 months.
- Add equal volume of > 95% ethanol directly to the homogenate cell sample. Mix thoroughly by vortexing.
- Library construction
- Qualify total RNA on Agilent 2100 Bioanalyzer using Agilent RNA 6000 Pico Kit. Purify polyA RNA from 50 ng total RNA using Oligo-dT Dynabeads according to manufacturer’s protocol (Dynabeads® mRNA DIRECTTM Purification Kit) and elute the polyA RNA in 12 μl RNase free water.
- RNA fragmentation
PolyA RNA 12 μl
5x first strand buffer 6 μl
Incubate at 95 °C 5 min and chill on ice - Reverse transcription
Add 1.5 μl Hexadeoxyribonucleotide mixture (50 ng/μl) to each sample, incubate at 65 °C for 5 mins and chill on ice.
Prepare the following RT mix on ice (1x):
dNTP mixture (10 mM) 1.5 μl
DTT (0.1 M) 3 μl
Ribonuclease inhibitor (40 U/μl) 1 μl
SuperScript II Reverse Transcriptase (200 U/μl) 1 μl
Nuclease-free water 4 μl
Add 10.5 µl of the RT mix to each sample, mix and spin. Transfer to thermocycler:
25 °C 10 min
42 °C 50 min
70 °C 15 min
4 °C hold - Synthesis second strand of cDNA
First strand cDNA 30 μl
dNTP mixture (10 mM) 2 μl
Second strand buffer (5x) 10 μl
RNase H (5 U/μl) 1 μl
DNA polymerase I (5 U/μl) 3 μl
Nuclease-free water 4 μl
16 °C 2.5 h
4 °C hold
Purify cDNA using 1.8x AMPure XP beads, and elute in Nuclease-free water. - Construct cDNA library using NEBNex® DNA library prep master mix set and NEBNext® multiplex oligos.
- Qualify library on Agilent 2100 Bioanalyzer using Agilent High Sensitivity DNA Kit. Quantify library using Qubit® dsDNA HS Assay Kit and Illumina Library Quantification Kit according to manufacturer’s protocol. Run library on the Illumina Hiseq-2500 sequencing system.
- Qualify total RNA on Agilent 2100 Bioanalyzer using Agilent RNA 6000 Pico Kit. Purify polyA RNA from 50 ng total RNA using Oligo-dT Dynabeads according to manufacturer’s protocol (Dynabeads® mRNA DIRECTTM Purification Kit) and elute the polyA RNA in 12 μl RNase free water.
Data analysis
- Mapping and analysis of Illumina reads for transcriptome
- Illumina Casava1.8 software used for basecalling.
- Drosophila melanogaster genome sequences and bowtie index can be downloaded from Tophat website (Figure 3).
Figure 3. Screenshots of the Tophat webpage - Single-end reads are mapped to the Drosophila melanogaster genome (Release 5) using TopHat (v2.0.10). For our analysis, we allow up to 2 mismatches and aligned with command ‘tophat --bowtie1 –N 2 --no-coverage-search –G genes.gtf genome sample.fastq’, where genes.gtf is the structure of genes and sample.fastq is the raw sequencing reads of a sample.
- Cuffquant is used for quantifying gene and transcript expression levels for a sample BAM file with command ‘cuffquant -o outputdir genes.gtf -p 15 sample_aligned/accepted_hits.bam’. This step generates .cxb files which contain gene and transcript expression levels.
- We use cuffnorm for computing normalized expression values (FPKM) of genes of all samples. csDendro and MDSplot are used for provide insight into the relationships between replicates and different conditions. This step makes sure the replicates samples are consistent and can be used for subsequent analysis (Figure 4).
- Differentially expressed genes are identified by cuffdiff and significant differentially expression genes were filtered with P value ≤ 0.05, and fold change ≥ 2.
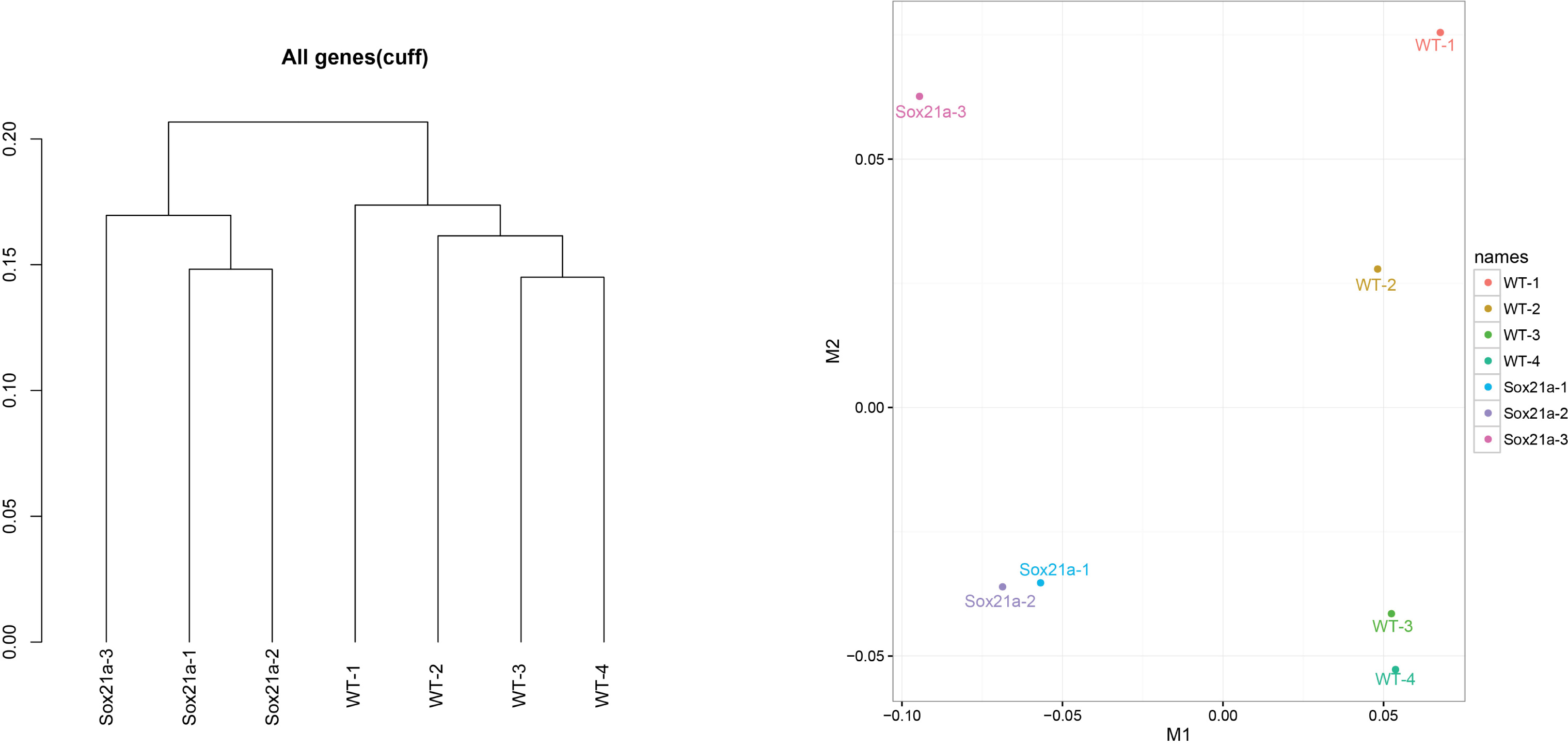
Figure 4. csDendro (left) and MDSplot (right) are used to evaluate the relationships among experimental and control groups. Here the example shows 3 replicates of sox21a mutant samples (Sox21a-1, -2, -3) and 4 replicates of WT control samples (WT-1, -2, -3, -4).
- Illumina Casava1.8 software used for basecalling.
Recipes
- 1x DEPC-PBS
1x PBS solution (pH 7.2-7.4) contains 137 mM NaCl, 2.7 mM KCI, 4.3 mM Na2HPO4 and 1.4 mM KH2PO4
0.1% final solution of diethyl pyrocarbonate (DEPC) is added to 1x PBS
Mix well by shaking and leave overnight in a fume hood at room temperature
The solution can be stored at room temperature up to a year in RNase-free conditions - Elastase solution
Dissolve elastase in 1x DEPC-PBS buffer at a final concentration of 1 mg/ml
Acknowledgments
Sorting of intestinal progenitor cells was performed according to the method described previously (Dutta et al., 2015) with some modifications. This work was supported by National Basic Science 973 grant (2014CB850002) from the Chinese Ministry of Science and Technology.
References
- Chen, J., Xu, N., Huang, H., Cai, T. and Xi, R. (2016). A feedback amplification loop between stem cells and their progeny promotes tissue regeneration and tumorigenesis. Elife 5. pii: e14330
- Dutta, D., Buchon, N., Xiang, J. and Edgar, B. A. (2015). Regional cell specific RNA expression profiling of FACS isolated Drosophila intestinal cell populations. Curr Protoc Stem Cell Biol 34: 2F 2 1-14.
- Langmead, B., Trapnell, C., Pop, M. and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3): R25.
- Trapnell, C., Pachter, L. and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9): 1105-1111.
- Roberts, A., Trapnell, C., Donaghey, J., Rinn, J. L. and Pachter, L. (2011). Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol 12(3): R22.
Article Information
Copyright
Chen et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Chen, J., Li, J., Huang, H. and Xi, R. (2016). Gene Expression Analysis of Sorted Cells by RNA-seq in Drosophila Intestine. Bio-protocol 6(24): e2079. DOI: 10.21769/BioProtoc.2079.
- Chen, J., Xu, N., Huang, H., Cai, T. and Xi, R. (2016). A feedback amplification loop between stem cells and their progeny promotes tissue regeneration and tumorigenesis. Elife 5. pii: e14330
Category
Stem Cell > Adult stem cell > Intestinal stem cell
Cell Biology > Cell isolation and culture > Cell isolation
Molecular Biology > RNA > RNA sequencing
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









