- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantitative 3D Time Lapse Imaging of Muscle Progenitors in Skeletal Muscle of Live Mice
Published: Vol 6, Iss 24, Dec 20, 2016 DOI: 10.21769/BioProtoc.2066 Views: 11339
Reviewed by: Antoine de MorreeXiaoyi ZhengAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Simplified 3D-Plasma Culture Method for Generating Minimally Manipulated Autologous Equine Muscle-Derived Progenitor Cells
Hélène Graide [...] Didier Serteyn
Dec 5, 2025 1251 Views

Monitoring of Sperm-Independent Calcium Oscillations in Immature Oocytes of Mice
Sae Horiike [...] Hidehiko Ogawa
Feb 5, 2026 63 Views

High Content In Vitro Survival Assay of Cortical Neurons
Paolo V. Fioretti [...] Manuela Basso
Feb 5, 2026 69 Views
Abstract
For non-optically clear mammalian tissues, it is now possible to use multi-photon microscopy to penetrate deep into the tissue and obtain detailed single cell images in a live animal, i.e., intravital imaging. This technique is in principle applicable to any fluorescently marked cell, and we have employed it to observe stem cells during the regenerative process. Stem cell-mediated skeletal muscle regeneration in the mouse model has been classically studied at specific time points by sacrificing the animal and harvesting the muscle tissue for downstream analyses. A method for direct visualization of muscle stem cells to gain real-time information over a long period in a live mammal has been lacking. Here we describe a step-by-step protocol adapted from Webster et al. (2016) to quantitatively measure the behaviors of fluorescently labeled (GFP, EYFP) muscle stem and progenitor cells during homeostasis as well as following muscle injury.
Keywords: Muscle stem cellBackground
Long-term in vivo imaging of stem and progenitor cells was first used for hair follicles during continuous physiological regeneration without surgical procedure (Rompolas et al., 2012). By contrast, stem cells for skeletal muscles are largely quiescent and inactive during the normal homeostatic state. An injury to the muscle is necessary to activate muscle stem cells to mount a regenerative process. In vitro live imaging of muscle stem/progenitor cells has been widely used to study them in artificial settings. To understand muscle stem cell behavior during regeneration in their native environment, we developed a method to image them during skeletal muscle regeneration. Our method allows up to 8 h of continuous imaging per session daily following injury. This is the first time that skeletal muscle stem cells have been observed in vivo in an injured/regenerative environment (Webster et al., 2016).
Materials and Reagents
- Lab tape (VWR, catalog number: 89097 )
- Razor blades for shaving hair (VWR, catalog number: 55411-060 )
- Kimwipes (ULINE)
- Transfer pipette (Globe Scientific, catalog number: 137038 )
- Cyanoacrylate based adhesive that bonds skin to glass (i.e., Loctite glass glue; Amazon.com; Scognamiglio et al., 2016)
- Mouse with GFP or YFP expression in Pax7-expressing muscle stem cells (or cell types of interest); GFP and YFP Cre-reporter mice and Pax7-Cre-ERT2 mouse strains are available at Jackson Laboratory (THE JACKSON LABORATORY, catalog numbers: 021847 , 006148, and 012476, respectively)
- Isoflurane (Patterson Veterinary Supply, IsoFlo®, catalog number: 07-806-3204 ; To be applied directly into the vapor chamber of equipment list 4; indicated by arrow in Figure 2)
- EtOH, 70% (Decon Labs, catalog number: V1401 )
- Distilled water (in house)
Equipment
- An inverted Leica SP5 (or equivalent) equipped with a Leica 25x/0.95 water objective, a 35 mm culture dish holder attached to the stage, and nondescanned detectors with a dichroic mirror separating the detectable spectrum (430-550 nm) at 495 nm (Figure 1)

Figure 1. Inverted Multi-photon setup. The imaging setup includes an inverted confocal microscope with 25x water objective fitted with a culture dish holder, multi-photon laser and computer. - Chameleon Vision laser (Coherent)
- Heating pad with enough area to cover the animal on the microscope stage (Sunbeam 756-500 Heating Pad from Amazon.com)
- Animal anesthesia system equipped with induction chamber as well as tubing with nose-cone (VetEquip, catalog number: 901806 ). Please consult with the operation manual prior to use
- 35 mm fluorodish (World Precision Instruments, catalog number: FD35-100 )
- Metal spatula (VWR, catalog number: 82027-530 )
- Heat protective glove (VWR)
- Bunsen burner (VWR, catalog number: 89038-528 )
- Laminar flow fume hood
- Fine tip forceps (Dumont #5 forceps) (Fine Science Tools, catalog number: 11252-30 )
- Fine scissors for cutting skin (straight, 11.5 cm) (Fine Science Tools, catalog number: 14058-11 )
- Spring scissors for cutting fascia (8 mm blades) (Fine Science Tools, catalog number: 15009-08 )
Software
- Leica SP5 software
- Fiji (Fiji is an updated version of ImageJ, an open source image processing software; Schindelin et al., 2012)
- Imaris (Bitplane, version 7.6.4 for Windows X64)
Procedure
Ethical statement: All procedures discussed here are in accordance with and were approved by the Carnegie Institution for Science Institutional Animal Care and Use Committee.
- Intravital imaging setup
- Turn on inverted confocal microscope and multi-photon (MP) laser. Set MP to 920 nm for GFP detection, or 940 nm for EYFP detection.
- Start Leica imaging software.
- Turn on heating pad and warm to 35 °C.
- Begin oxygen flow to anesthesia machine and then begin filling induction chamber with 1.7% isoflurane (by adjusting the turn dial on top of the container to 1.7) and the flow-meter indicator ball to 1 (for 1 L/min flow rate) (See Figure 2).
- Place mouse in induction chamber to begin anesthetization (Figure 2) and continue onto making modified fluorodish. The anesthetized mouse can stay in the induction chamber for this duration (~20 min). If the fluorodish preparation takes longer, the mouse can be left in the induction chamber without concern, as they will continue to be placed under similar anesthesia condition during imaging.
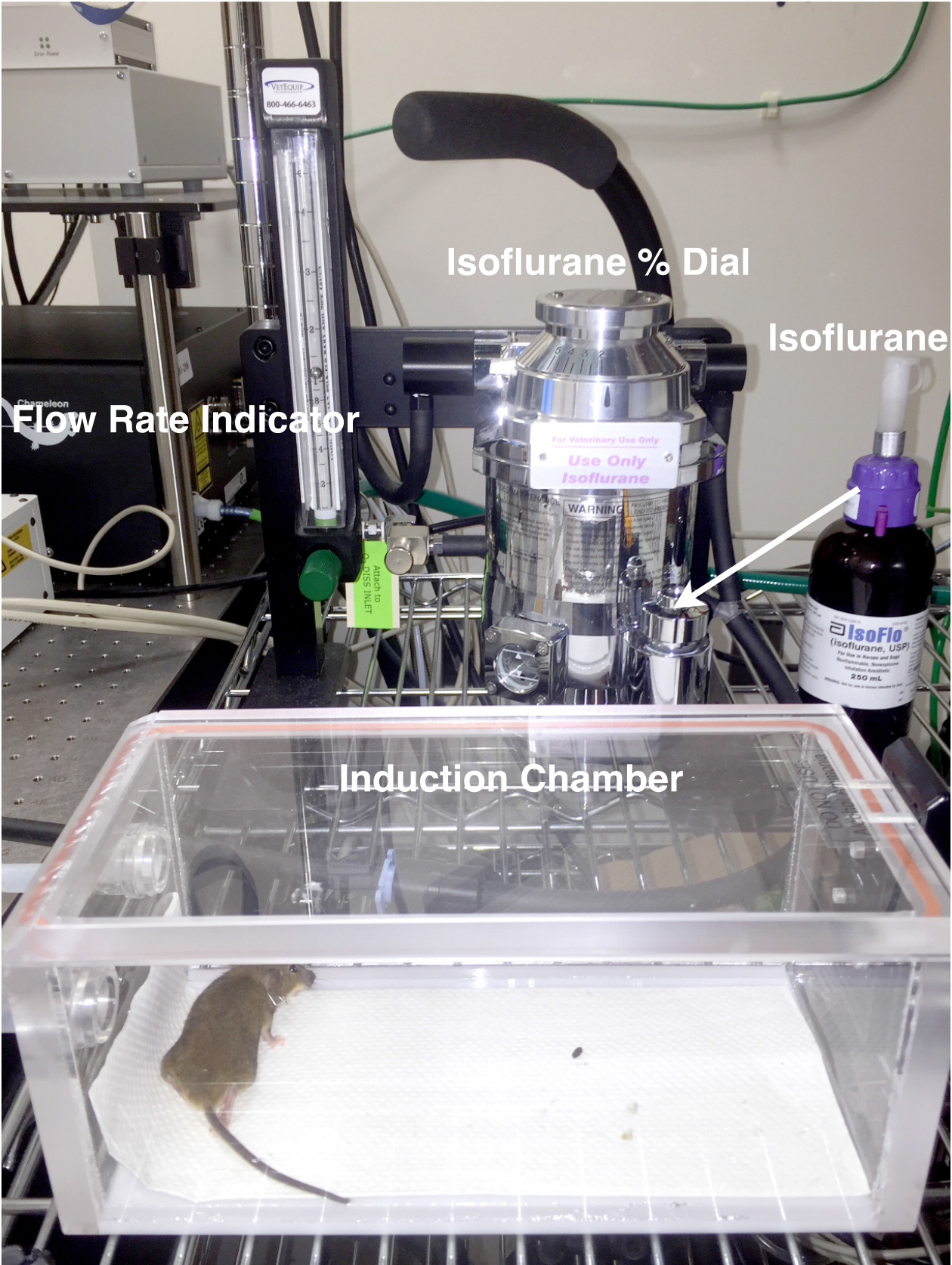
Figure 2. Anesthetization setup. This consists of anesthesia machine with regulator, induction chamber and nose-cone (pictured in Figure 4). - Prepare modified fluorodish for mounting on mouse’s leg
- Wearing a heat protective glove, heat metal spatula over Bunsen burner in a fume hood.
- Use heated spatula to melt and remove a 2-3 cm section of the fluorodish wall. Take care not to melt and deform the bottom of the fluorodish, which can be problematic when fitting the fluorodish into the microscope stage holder (see step 4e). It is important to leave part of the fluorodish to secure and stabilize it in the stage holder.
- Repeat steps 2a and 2b and remove a second section from the opposite side of the fluorodish.
- This can be done ahead of time; multiple fluorodishes can be prepared and stored for future use (Figure 3).
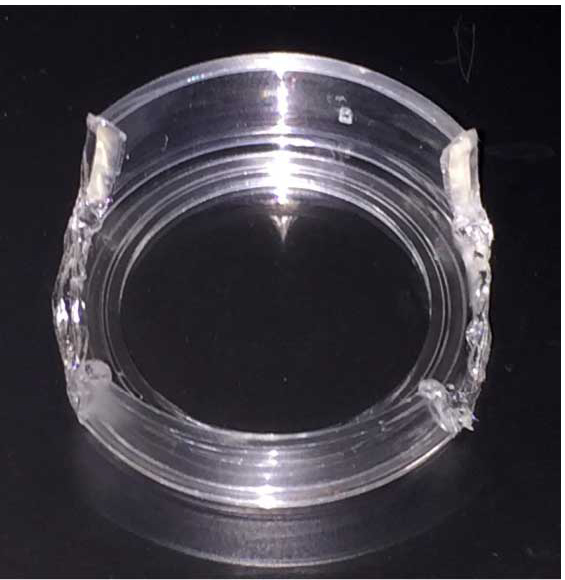
Figure 3. Modified fluorodish
- Wearing a heat protective glove, heat metal spatula over Bunsen burner in a fume hood.
- Mount fluorodish on mouse
- Move mouse from induction chamber to procedure surface where anesthesia tubing with attached nose-cone has been fastened with tape (Figure 4).
- Secure mouse’s head by placing a small piece of tape over the head across to the nose-cone.
- Check level of anesthetization by observing the mouse. The mouse should maintain a steady rate of respiration but not react to slight pinches with forceps on the skin of the leg. If the mouse’s respiration is very slow, decrease the Isoflurane to 1.3% to 1.5%. If the mouse flinches when leg is pinched, then wait longer for mouse to enter deeper state of anesthetization before proceeding.
- Use a razor blade to shave hair from skin covering the tibialis anterior (TA) muscle. Begin shaving at the ankle and work up to the knee. Remove any lose hair from the lower leg. This can be done by washing the skin with 70% EtOH followed by Kimwipes to remove loose hairs.
- Ready adhesive and set aside.
- Cut a thin strip of skin from the region over the TA by pulling a small piece of skin up with forceps and cutting with scissors, beginning near the ankle and cutting up towards the knee. Do not remove too much skin, only enough to expose a 1-2 cm2 area over the TA.
- Remove the fascia covering the TA by gently pulling up on the membrane at the side of the TA and then cutting with scissors. One may avoid damaging the muscle by using forceps to pull the membrane at the edge of the TA without making direct contact with tissue that is to be imaged.
- Place the modified fluorodish on the mouse’s leg so that the surface of the dish is flush with the exposed muscle and the mouse’s upper leg/body and lower leg/ankle fit in the spaces in the dish wall that have been removed. Secure the dish in place by holding the mouse’s leg in contact with the glass bottom of the dish. The remaining sides of the modified fluorodish should run parallel with the length of the mouse’s limb to be imaged.
- While holding the dish in place, apply adhesive where the skin of the leg is in contact with the glass. Apply adhesive around the entire perimeter and let the adhesive set for 90 sec or more while holding the dish in place.
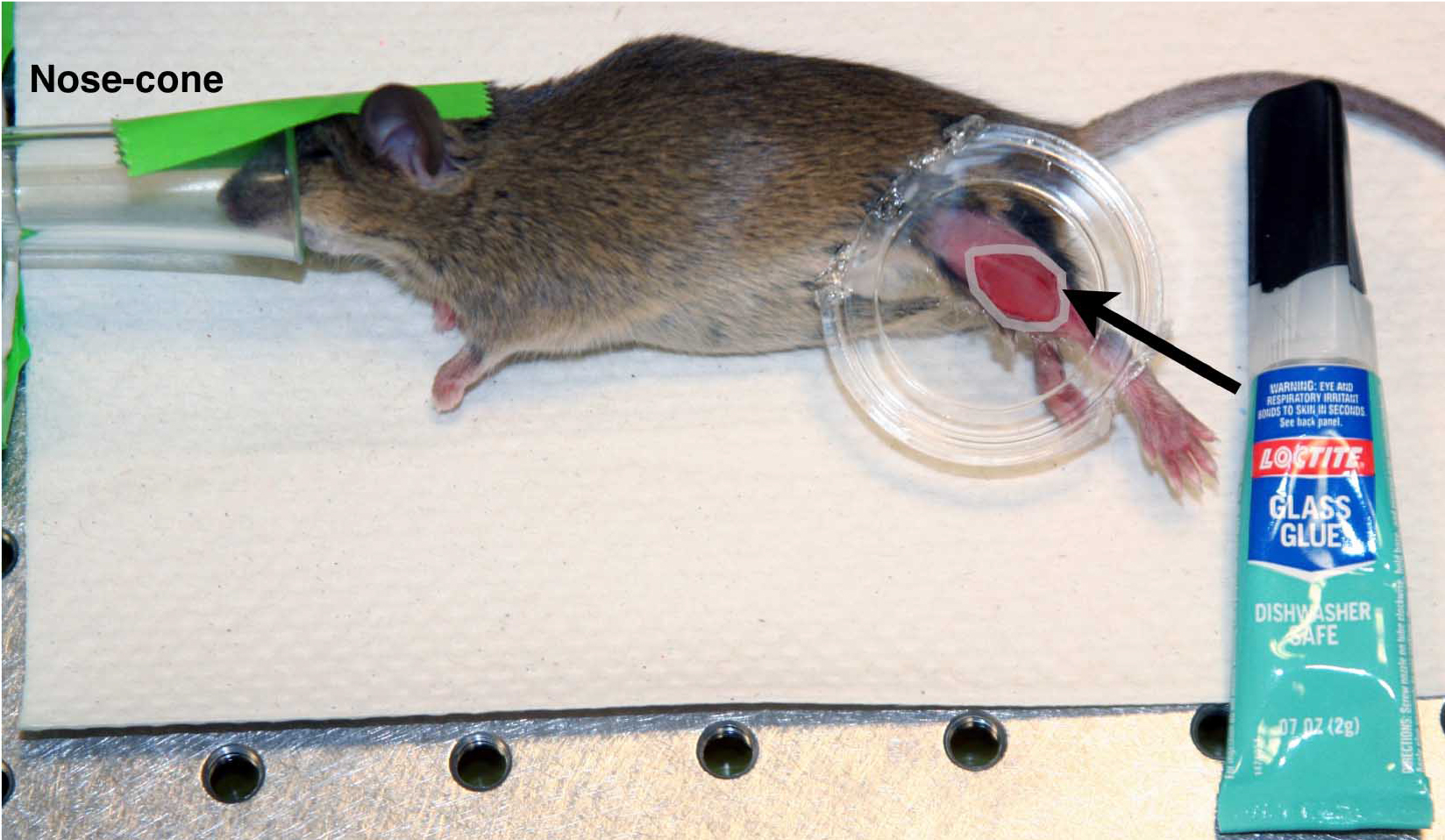
Figure 4. Fluorodish mounted on mouse. Pseudo-colored white ring indicates position to apply adhesive, performed using nose-cone anesthesia. - Begin filling the induction chamber with 1.7% isoflurane at 1 L/min. Maintain flow to the nose-cone.
- Once adhesive has set, return mouse to induction chamber and stop flow to the nose-cone.
- Position mouse for intravital imaging
- Tape the anesthesia tube with nose-cone to the microscope stage.
- If using a water immersion objective, apply water to the surface of the objective.
- Switch delivery of 1.7% isoflurane at 1 L/min to the nose-cone outlet.
- Remove mouse from induction chamber and place on microscope stage, placing the head of the mouse into the nose-cone. Secure the head to the nose-cone with tape. The mouse should be positioned on its abdomen. When placed on its back, lungs often collapse prematurely terminating imaging (~1-2 h).
- Place the fluorodish in the culture dish holder affixed to the stage (Figure 5I).
- Raise the objective lens into position and identify the imaging field using the epifluorescence function of the Leica SP5. A piece of tape may be placed across the lower portion of the mouse’s body (attached on either side to the stage) to reduce movement from respiration.
- Place heating pad over mouse (Figure 5II).
- Reduce anesthesia to 1.3% isoflurane at 1 L/min.

Figure 5. Mouse positioned for intravital imaging. I. Stage setup. A) Exposed TA muscle for imaging mounted to modified fluorodish. B) Nose-cone anesthesia apparatus adhered to stage. C) Culture dish holder. D) 25x water objective with H2O (pictured below field of view). II. Microscope setup with positioned heating pad. - Image capture parameters and analyses
- Capture serial optical sections at 3 μm steps to a total depth of 150-200 μm every 3 min for up to 8 h. Scan fields of 0.31 to 0.62 mm2 at 400 Hz. Following imaging, the mouse is sacrificed. Due to the adhesive used, it is not possible to remove fluorodish without causing additional tissue damage for proper suture, and the fluorodish is too bulky to be left on the mouse without affecting mobility.
- Image sequence files (.lif) can be opened in Fiji and converted to .tiff stacks or image sequences for analysis.
Data analysis
Data storage, transfer, and processing:
- A Leica generated .lif file can range from 3 to 8 GB.
- A single .tiff image sequence (converted from .lif using Fiji) is about 3 GB (3D time lapse).
- A single .tiff stack (3D time lapse) is ≤ 1 GB.
- A single .tiff stack (2D time lapse) is ≤ 100 MB.
- An .avi movie is ≤ 20 MB.
- Image sequences can be aligned using the Fiji plugin, ImageJ 2.0.0 ‘Correct 3D drift’.
- Drift-corrected image sequences can be analyzed further with Fiji (e.g., long axes and division angle measurements of cells, as well as measurements of cell migration direction). Imaris 7.6.4 is used for 4D rendering, cell tracking, migration statistics, and 4D movies.
- The processed data (images and videos) and statistical analyses are published in Webster et al. (2016), which can be found at: http://www.cell.com/cell-stem-cell/fulltext/S1934-5909(15)00503-2
Notes
- Obstructions to light transmission include hair, red blood cells, the epimysium, and adhesive. These can be avoided by removing all lose hair from the leg during step 3d, minimizing skin removal and damage to vasculature during steps 3e and 3f, removing all membrane covering the exposed TA in step 3f, and ensuring that adhesive does not cover the exposed tissue in step 3i.
- Mounting the fluorodish on the leg can be quite challenging and may require some practice. Fitting a P1000 pipette tip on the adhesive tube can allow for easier access and delivery of adhesive to the interface of skin and glass at the perimeter of the exposed TA.
- A shroud placed over the stage/microscope may be necessary to block ambient light from causing high background signal, especially when using very sensitive non-descanned detectors (Figure 6).
- For long duration imaging sessions, repeated applications of water are needed. To overcome the hassle of pausing the experiment and applying water to the objective lens, a water delivery objective lens ring cover and pump apparatus can be used (New Era Pump Systems, Inc.).
- This protocol can be adapted to other fluorescent proteins (i.e., tdT, CFP) by adjusting MP wavelength and detection filters accordingly.
- All reagents used in this protocol were purchased and used directly without any dilution or mixing.

Figure 6. Imaging with shroud
Acknowledgments
The protocol reported here was supported by the NIAMS of the NIH under award numbers F32AR065366 (to MTW) and RO1AR060042 (to C-MF), as well as by intramural funds of the Carnegie Institution (to TH). This protocol is a modified version derived from a previous publication in Cell Stem Cell (cited in the reference, Webster et al., 2016).
References
- Rompolas, P., Deschene, E. R., Zito, G., Gonzalez, D. G., Saotome, I., Haberman, A. M. and Greco, V. (2012). Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487: 496-499.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P. and Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682.
- Scognamiglio, F., Travan, A., Rustighi, I., Tarchi, P., Palmisano, S., Marsich, E., Borgogna, M., Donati, I., de Manzini, N. and Paoletti, S. (2016). Adhesive and sealant interfaces for general surgery applications. J Biomed Mater Res B Appl Biomater 104(3): 626-639.
- Webster, M. T., Manor, U., Lippincott-Schwartz, J. and Fan, C. M. (2016). Intravital imaging reveals ghost fibers as architectural units guiding myogenic progenitors during regeneration. Cell Stem Cell 18(2): 243-252.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Webster, M. T., Harvey, T. and Fan, C. (2016). Quantitative 3D Time Lapse Imaging of Muscle Progenitors in Skeletal Muscle of Live Mice. Bio-protocol 6(24): e2066. DOI: 10.21769/BioProtoc.2066.
Category
Stem Cell > Adult stem cell > Muscle stem cell
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











