- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Shoot Apical Meristem Size Measurement
Published: Vol 6, Iss 23, Dec 5, 2016 DOI: 10.21769/BioProtoc.2055 Views: 9944
Reviewed by: Tie LiuYuan ChenAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Modified Pseudo-Schiff Propidium Iodide for Staining the Shoot Apical Meristem in Arabidopsis
Ruiqi Li [...] Ligeng Ma
May 5, 2023 2269 Views

A Novel Imaging Protocol for Investigating Arabidopsis thaliana Siliques and Seeds Using X-rays
Brylie A. Ritchie [...] Ansul Lokdarshi
Oct 5, 2023 2183 Views

Direct Plant Regeneration From Immature Male Inflorescence of Banana (Musa spp.)
Pradeep Chand Deo
Oct 20, 2025 1426 Views
Abstract
The shoot apical meristem (SAM) is a collection of cells that continuously renew themselves by cell division and also provide cells to newly developing organs. It has been known that CLAVATA (CLV) 3 peptide regulates a transcription factor WUSCHEL (WUS) to keep numbers of undifferentiated cells constant and maintain the size of the SAM. The interactive feedback control of CLV3 and WUS in a non-cell autonomous signaling cascade determines stem cell fate (maintenance of pluripotency or, alternatively, differentiation into daughter cells) in the SAM. Ca2+ is a secondary messenger that plays a significant role in numerous signaling pathways. The signaling system connecting CLV3 binding to its receptor and WUS expression is not well delineated. We showed that Ca2+ is involved in CLV3 regulation of the SAM size. One of the approaches we used was measuring the size of the SAM. Here we provide a detailed protocol on how to measure Arabidopsis SAM size with Nomarski microscopy. The area of the two-dimensional dome representing the maximal ‘face’ of the SAM was used as a proxy for SAM size. Studies were done on wild type (WT) Arabidopsis in the presence and absence of a Ca2+ channel blocker Gd3+ and the CLV3 peptide, as well on genotypes that lack functional CLV3 (clv3) or a gene encoding a Ca2+-conducting ion channel (‘dnd1’).
Keywords: ArabidopsisBackground
Nomarski microscopy is widely used to study Arabidopsis SAM size. Other microscopy techniques for SAM observation are time consuming and require embedding tissue in resin and then sectioning or even more sophisticated microscopy. Nomarski microscopy, along with tissue clearing techniques is fast and convenient for whole tissue imaging. Published methods on SAM size measurement with Nomarski microscopy are often briefly described. Here, we provide a modified protocol with a detailed step by step guide including steps from dissecting Arabidopsis SAM tissues, through sample preparation for Nomarski microscopy, and SAM size measurement.
Materials and Reagents
- 3 x 4 cell culture multi-well plates (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 150628 )
- Parafilm
- Razor blade
- Glass slide (75 x 38 mm) (Thermo Fisher Scientific, Fisher Scientific, catalog number: 12-550B )
- Coverslips (18 x 18 mm) (thickness: 0.17 mm) (Thermo Fisher Scientific, Fisher Scientific, catalog number: S17521 )
- Arabidopsis thaliana wild type (ecotype Columbia), dnd1 mutant (At5g15410), clv3 mutant (At2g27250)
- Synthetic CLV3 peptide RTVPhSGPhDPLHH3 (GenScript, Piscataway, NJ)
- Murashige and Skoog salts (MS) (Caisson Laboratories, catalog number: MSP01-10LT )
- Gadolinium(III) chloride (GdCl3) (Sigma-Aldrich, catalog number: 439770 )
- Ethanol (Sigma-Aldrich, catalog number: 459836 )
- Sucrose (Sigma-Aldrich, catalog number: S0389 )
- MES buffer (pH 5.7) (Caisson Laboratories, catalog number: M009-100GM )
- Sterilized water
- Tris (Sigma-Aldrich, catalog number: 252859 )
- Acetic acid (Thermo Fisher Scientific, Fisher Scientific, catalog number: A38S-500 )
- Chloral hydrate (Sigma-Aldrich, catalog number: C8383 )
- Glycerol (Thermo Fisher Scientific, catalog number: 17904 )
- Plant culture medium (see Recipes)
- Fixing solution (see Recipes)
- Clearing solution (see Recipes)
Equipment
- Fridge
- Shaker
- Growth chamber for growing plants (100 µmol m-2 sec-1 white light for 16 h and dark for 8 h, 23 °C)
- Tweezers
- Dissecting microscope
- Microscope equipped with Nomarski optics (Nikon Instrument, model: MICROPHOT-FX )
Software
- Infinity analyze program (Lumenera, Ottawa, Canada)
Procedure
- Arabidopsis seedling culture and treatments
- Add 2 ml ½ MS liquid media to each well of a multi-well plate.
- Surface sterilize all seeds and place 1 seed into each well of the multi-well plate.
- Stratify the seeds by placing the multi-well plate at 4 °C for 3 days to break dormancy.
- After stratification, take the multi-well plate out from the 4 °C fridge and add different treatments to each well. In our experiment, we added CLV3 ligand alone (1 μM final concentration), Ca2+ channel blocker GdCl3 alone (150 μM final concentration) and CLV3 with GdCl3 together (at final concentrations of 1 and 150 μM, respectively) to ½ MS liquid media.
Note: Because CLV3 peptide was dissolved in water, the control group should be water and should be the same amount as the CLV3 treatment group. The small amount of water added (in our experiment was 2 μl) should not have any osmotic stress on seedlings. - Put the multi-well plate containing seedlings on a rotatory shaker (180 rpm) in a growth chamber (100 µmol m-2 sec-1 white light for 16 h and dark for 8 h, 23 °C). We define this day as our beginning day of experiment (day 0).
- Add 2 ml ½ MS liquid media to each well of a multi-well plate.
- Seedling preparation for dissection
- After 7 days of growth in the growth chamber, gently take out individual seedlings one at a time from the multi-well plate with a tweezer and put it on a piece of paper towel for 3 sec to absorb excessive media.
- Remove the individual seedling from the paper towel with a tweezer and lay it sideways on a piece of Parafilm under a dissecting microscope.
- Remove the roots, cotyledons and the oldest leaf primordia with a sharp razor blade.
- To prevent morphology change, quickly (but gently) move dissected tissue with a tweezer into a well of the same plate containing only freshly made fixing solution and incubate overnight at room temperature.
- Remove fixing solution the next day and replace with 90% ethanol. Put the plate on a shaker at room temperature for up to 1 h (40 rpm).
- Remove 90% ethanol and replace with 70% ethanol. Put the plate on a shaker at room temperature for up to 1 h (40 rpm).
- Remove dissected tissue one at a time from the 70% ethanol and place on a glass slide with a drop of freshly made clearing solution.
- Observe under a microscope equipped with Nomarski optics (MICROPHOT-FXA; Nikon, Tokyo, Japan).
Note: The reaction of clearing solution is fast (~5 min). It is best to look at 1 to 2 samples at a time and take pictures as soon as the SAM is observed under the microscope. After a while the dome of the SAM will be hard to distinguish from surrounding tissues. The rest of the seedlings can be kept in 70% ethanol before observing under microscope.
- After 7 days of growth in the growth chamber, gently take out individual seedlings one at a time from the multi-well plate with a tweezer and put it on a piece of paper towel for 3 sec to absorb excessive media.
- SAM area measurement.
- Open Infinity analyze software program.
- Select ‘capture’ to take photographs. Record individual seedling images at the focal plane corresponding to the median optical section of the SAM.
- Draw a straight line between the basal edges of two opposing leaf primordia on each side of the SAM dome. This line represents the base of the SAM dome (Figure 1). A perpendicular line was generated connecting the midpoint of the SAM base to the top of the SAM dome (Figure 1C).
- Select ‘Area perimeter’ for SAM size measurement. Circle a closed SAM shape area based on the two lines described in step C3 by freehand drawing. The area of the SAM is determined by calculating the area above the straight line that represented the width of the base of the SAM. The program will automatically calculate the area and the perimeter.

Figure 1. 7-d-old Arabidopsis SAM area under a microscope equipped with Nomarski optics. A. Wild type. B. DEFENSE NO DEATH 1 (dnd1) mutant, which lacks a functional cyclic nucleotide gated cation channel isoform 2 (CNGC2) polypeptide and (C) clv3 mutant. Yellow triangles indicate the basal edges of the SAM dome. The red line that connects the 2 yellow triangles represents the base of the SAM dome. The perpendicular red line connects the midpoint of the SAM base to the top of the SAM dome.
- Open Infinity analyze software program.
Data analysis
For comparison between SAM size of wild type, dnd1 and clv3 mutant seedlings (see Figure 1 and Figure 2A), at least 15 seedlings of each genotype were measured. For calcium channel blocker effects on endogenous CLV3 in wild type (Figure 2B) and exogenous CLV3 effects on clv3 mutant, at least 10 seedlings were measured for each treatment (Figure 2C). ANOVA analysis was used to evaluate means separation. For Figures 2A and 2B, an asterisk or two asterisks above the bar representing a genotype or treatment indicate SAM size was significantly different (at P < 0.05 or P < 0.01, respectively) than control (WT). For Figure 2C, ANOVA comparisons are indicated by brackets.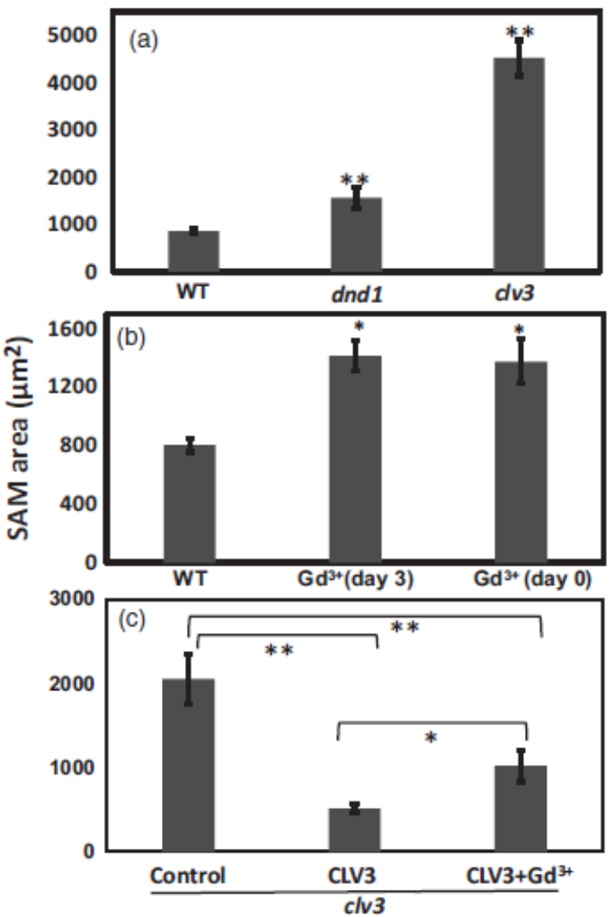
Figure 2. Ca2+ signaling interacts with CLV3 control of SAM area. (a). SAM area of 7-d-old WT, dnd1 and clv3 seedlings. (b). WT seedlings grown on standard ½ MS liquid medium (Control), or medium supplemented with Gd3+ µM on day 3 or on day 0 (the day the experiment starts). (c). clv3 seedlings grown on standard medium were treated with water (control), CLV3, or CLV3 and Gd3+. CLV3 and Gd3+ were applied on day 0. ANOVA analysis was used to evaluate means separation. All seedlings shown in Figure 2 are 7-d-old seedlings. For (a) and (b), an asterisk or two asterisks above the bar representing a genotype or treatment indicate SAM size was significantly different (at P < 0.05 or P < 0.01, respectively) than control (WT). For (c), ANOVA comparisons are indicated by brackets.
Recipes
- Plant culture medium
Dissolve 2 g sucrose, 0.433 g ½ MS salt, and 2 ml MES in 200 ml MiliQ water, and then adjust the pH to 5.7 with Tris. Autoclave medium - Fixing solution
Acetic acid:absolute ethanol (1:9, v/v) - Clearing solution
Chloral hydrate, glycerol and water (8:1:2, w/v/v)
Acknowledgments
This work was supported by National Science Foundation award 1146827 (to G.A.B). This protocol was adapted and modified from Fiers et al., (2006), Ohyama et al., (2009), Carles et al., (2010) and Dr. Miguel Aguilar.
References
- Aguilar M. Identifying features of mutant seeds using nomarski microscopy (Gene one).
- Carles, C. C., Ha, C. M., Jun, J. H., Fiume, E. and Fletcher. J. C. (2010). Analyzing shoot apical meristem development. Plant Development Biol 655:105-29.
- Fiers, M., Golemiec, E., Schors, R., Van, Der., Geest, L., Van, D., Li, K. W., Stiekema, W. J. and Liu, C. M. (2006). The CLAVATA3 / ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol 141(4): 1284-1292.
- Ohyama, K., Shinohara, H., Ogawa-Ohnishi, M. and Matsubayashi, Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol 5(8): 578-580.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chou, H., Wang, H. and Berkowitz, G. A. (2016). Shoot Apical Meristem Size Measurement. Bio-protocol 6(23): e2055. DOI: 10.21769/BioProtoc.2055.
Category
Plant Science > Plant developmental biology > Morphogenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









