- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vivo Imaging of Tumor and Immune Cell Interactions in the Lung
Published: Vol 6, Iss 20, Oct 20, 2016 DOI: 10.21769/BioProtoc.1973 Views: 10736
Reviewed by: Kristopher MarjonAlka MehraElizabeth V. Clarke

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Lipid-Mediated Sequential Recruitment of Proteins Via Dual SLIPT and Dual SLIPTNVOC in Live Cells
Kristina V. Bayer and Richard Wombacher
Nov 5, 2025 1600 Views

Monitoring of Sperm-Independent Calcium Oscillations in Immature Oocytes of Mice
Sae Horiike [...] Hidehiko Ogawa
Feb 5, 2026 63 Views

High Content In Vitro Survival Assay of Cortical Neurons
Paolo V. Fioretti [...] Manuela Basso
Feb 5, 2026 64 Views
Abstract
Immunotherapy has demonstrated great therapeutic potential by activating the immune system to fight cancer. However, little is known about the specific dynamics of interactions that occur between tumor and immune cells. In this protocol we describe a novel method to visualize the interaction of tumor and immune cells in the lung of live mice, which can be applied to other organs. In this protocol fluorescent-labeled tumor cells are transferred to recipient mice expressing fluorescently tagged immune cells. Tumor-immune cell interactions in the lung are then imaged by confocal or two photon microscopy. Analysis of tumor interactions with immune cells using this protocol should aid in a better understanding of the importance of these interactions and their role in developing immunotherapies.
Keywords: In vivo imagingBackground
A number of immunotherapies have demonstrated great promise in treating cancer. Understanding the spatial temporal resolution of how these tumor-immune interactions occur is important for enhancing and developing new immunotherapies. In this protocol we describe a novel method to directly visualize tumor-immune cell interactions in vivo in mouse lung. This protocol is initially described in our work examining the interactions of patrolling monocytes and tumor cells in the mouse lung (Hanna et al., 2015). This fluorescent microscopy protocol uses the vacuum imaging ring to stabilize and image the lung, which was initially described by Looney and colleagues (Thornton et al., 2012). In this protocol fluorescent-labeled tumor cells are transferred to recipient mice expressing fluorescently tagged immune cells. Tumor-immune cell interactions in the lung are then imaged by confocal or two photon fluorescent microscopy using the vacuum imaging ring. This protocol allows for the addition of other immune cell markers by intravenous (IV) injection of fluorescently labeled antibodies, and is adaptable to image tumor-immune cell interactions in other organs. Quantitative information such as the localization, engulfment of tumor material, timing, speed and frequency of these immune cell interactions can be collected using this protocol. This protocol should aid in helping to better understand the specific immune-tumor cell interactions that are important to developing better immunotherapies in the future.
Materials and Reagents
- Cell culture flask
- 30 gauge insulin syringe (BD, catalog number: 328431 ) Nr4a1-GFP (The Jackson Laboratory, catalog number: 018974 ), CX3CR1-GFP (The Jackson Laboratory, catalog number: 005582 ) or other fluorescent reporter mouse for visualizing immune cells.
- ½ micro-cover glass (12 mm diameter) (Electron Microscope Sciences, catalog number: 72230-01 )
- PE-90 tubing (BD, IntramedicTM, catalog number: 427420 )
- Mice
- Lewis lung carcinoma cells expressing red fluorescent protein (LLC-RFP) or other fluorescent-tagged tumor cell line (AntiCancer.com)
- TrypLETM Express enzyme(Thermo Fisher Scientific, GibcoTM, catalog number: 12604013 )
- Dulbecco's phosphate-buffered saline (DPBS) (GE Healthcare, HycloneTM, catalog number: SH30038.02 )
- Ketamine hydrochloride
- Xylazine hydrochloride
- Vetbond glue (3M, catalog number: 1469SB )
- Oxygen
- Ethanol
- Dow Corning® high vacuum grease (Sigma-Aldrich, catalog number: Z273554 )
Equipment
- Centrifuge
- Mechanical mouse ventilator (Harvard Apparatus, model: 845 )
- Fine dissecting tweezers and scissors (Fine Scientific Tools)
- Suction ring for imaging (Mekilect, catalog number: Suction Ring )
- Vacuum line with pressure regulator and pressure gauge
- Upright confocal or 2photon microscope with resonance scanner (we use a Leica SP5), heated stage and long distance 20-25x water immersion objective suitable for live imaging.
Software
- Imaris software (version 7.1.1 x 64) (Bitplane, http://www.bitplane.com/download/manuals/ReferenceManual6_1_0.pdf)
- Prism software (GraphPad Software)
Procedure
- Tumor injection
- Collect LLC-RFP tumor cells at about 70-80% confluency grown in a cell culture flask. Wash with DPBS two times and then gently trypsinize cells for approximately 3-5 min. We find that the greatest amount of reproducibility with tumors that are 70-80% confluent and highly viable (> 95%) when collected for injection.
Note: If there is any question of tumor cell viability, test for viability by trypan blue staining or other method after tumor cell collection. - Resuspend cells in DPBS and centrifuge for 5 min at 300 x g. Repeat DPBS wash.
- Count cells and confirm high viability (> 95%), then resuspend cells in DPBS at 3 x 106 cells/ml.
- Intravenously inject (in tail vein) 3 x 105 Lewis lung carcinoma cells expressing red fluorescent protein (LLC-RFP) or other fluorescent-tagged tumor cell line resuspended in 100 μl of DPBS into the tail vein using a 30 gauge syringe.
- Collect LLC-RFP tumor cells at about 70-80% confluency grown in a cell culture flask. Wash with DPBS two times and then gently trypsinize cells for approximately 3-5 min. We find that the greatest amount of reproducibility with tumors that are 70-80% confluent and highly viable (> 95%) when collected for injection.
- Mouse imaging
- Inject mouse intraperitoneally with an initial dose of 90 mg/kg ketamine hydrochloride and 15 mg/kg xylazine hydrochloride in 0.5 ml of PBS to anesthetize mouse. Maintain anesthesia during image acquisition with one-half dose subcutaneously boosting every 45 min for up to 3 h.
- Placed mouse in the supine position on a heated microscope stage to achieve a rectal temperature of 37 °C.
- Once adequate anesthesia is induced (tested by paw pinch), expose the trachea and separate from the surrounding connective tissue.
- Cut a small opening in the anterior trachea parallel to the cartilaginous rings, and insert PE-90 tubing trimmed at a 45 degree angle into the trachea toward the lungs. Secure the tubing and close the skin around the trachea with Vetbond glue and connect to the mechanical ventilator. Apply a tidal volume of 8 to 10 μl/g body weight (room air or higher fractions of oxygen) with respiratory rates of approximately 120 breaths per min. This is the approximate normal breathing rate of an average size adult mouse (25-30 g).
- Reassess depth of anesthesia with a paw pinch.
- Dab mouse lightly with 70% ethanol around left rib cage and remove skin to expose ribs.
- Gently insert scissors between the two lowest ribs and cut a 0.5 cm hole into the thoracic cavity.
- Pull up second rib above hole with tweezers and then pipette approximately 300 μl of PBS into hole to help drop left lobe of the lung from the rib cage.
- Continue applying upward pressure on the ribcage with tweezers in order to keep the lung away from the ribs and prevent lung damage while removing ribs. Carefully remove approximately 1.5 cm of the three anterior ribs overlying the left lung lobe as shown in Figure 1.
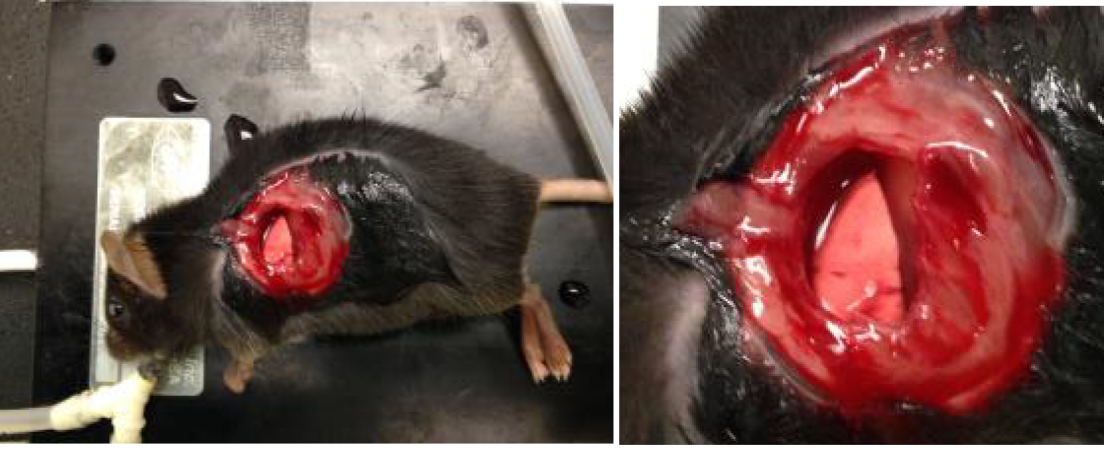
Figure 1. Images of exposure of left lobe of lung as described in step B9 - Apply a thin layer of high vacuum grease around the inner edge of the vacuum ring and then apply a 12-mm glass coverslip sealed with high vacuum grease to the vacuum suction ring. Apply 20 to 25 mmHg vacuum suction to the ring using a dedicated vacuum line attached to pressure gauge and regulator to adjust pressure.
- Carefully lower thoracic suction window onto the flat surface of the left lung lobe, which will then enter the thoracic window and be stabilized for imaging as shown in Figure 2.
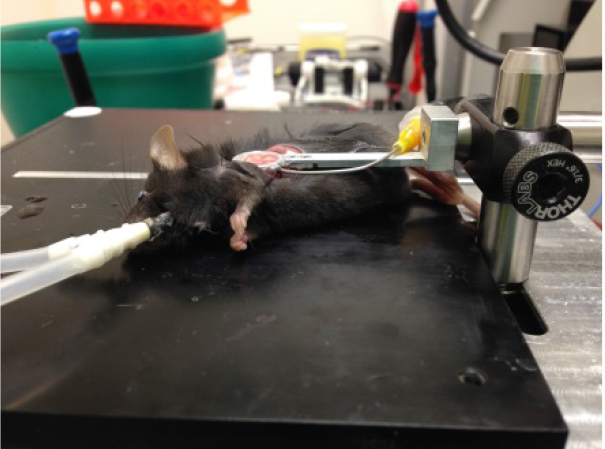
Figure 2. Image of suction window on left lobe of the lung - Add ~200 μl DPBS to slideglass, engage microscope lens on imaging ring as shown in Figure 3, and acquire imaging as necessary. Confirm normal blood flow in lung by imaging to ensure that the ring has not caused vascular damage/blocking. Focusing on imaging an area that is fairly stabilized by the vacuum ring to prevent motion artifact and distortion of the images by pulsing of the lung. Representative lung imaging is shown in Video 1 and Video 2.
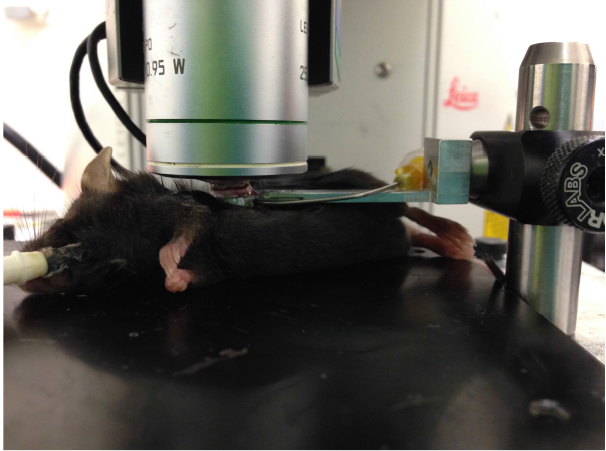
Figure 3. Image of microscope lens on lung suction ring connected to lung of live mouse
- After imaging turn off vacuum, remove microscope lens and imaging ring, and euthanize mouse.
Video 1. Representative live imaging of Nur77-GFP high patrolling monocytes (Green), all CD45-APC+ immune cells (Blue), and LLC-RFP tumor (Red) in mouse lung. Nur77-GFP mice were IV injected with LLC-RFP tumor 24 h prior to imaging. CD45-APC was IV injected.
Video 2. Representative three dimensional reconstruction of live imaging data using Imaris. Nur77-GFPhigh patrolling monocytes (Green), GR1(Ly6G/C)-APC+ neutrophils and classical monocytes (Blue), and LLC-RFP tumor (Red) in mouse lung. Nur77-GFP mice were IV injected with LLC-RFP tumor 7 days prior to imaging. GR1(Ly6G/C)-APC was IV injected.
- Inject mouse intraperitoneally with an initial dose of 90 mg/kg ketamine hydrochloride and 15 mg/kg xylazine hydrochloride in 0.5 ml of PBS to anesthetize mouse. Maintain anesthesia during image acquisition with one-half dose subcutaneously boosting every 45 min for up to 3 h.
Data analysis
Three dimensional reconstructions and drift correction of lung images were performed using Imaris software (version 7.1.1 x 64). Detailed instructions on the use of Imaris software can be found at: http://www.bitplane.com/download/manuals/ReferenceManual6_1_0.pdf. Images were smoothed by median filtering at kernel size 3 x 3 pixels. When needed, motion artifacts caused by breathing were corrected with rigid body registration using an ImageJ plug-in (McArdle et al., 2014; McArdle et al., 2015). Imaris software was also used to automatically process 3D video data by detecting cells in each fluorescence channel, then creating tracks by linking the detected cells over time (using the Imaris 'Track spots over time' function). Tracks were manually edited to improve accuracy. Data for all experiments were analyzed with Prism software. Unpaired t-tests and two-way analysis of variance were used for comparison of experimental groups. P values of less than 0.05 were considered significant. The data appeared to be normally distributed with similar standard deviation and error observed between and within experimental groups.
Notes
- Optional: Intravenously inject (retro-orbital) directly conjugated fluorescent antibodies (20 μg diluted to 100 μl in DPBS of either Ly6G: Neutrophils, CD3e: T Cells, CD31: endothelial cells, etc.) to label other immune cells or vasculature. Optimal color for antibody labeling will vary with microscope set-up but we find good imaging with APC/A647, FITC/A488, BV421 or PE labeling of antibodies. Antibodies should be injected just prior or during imaging. Good antibody labeling of cells can be detected in vivo for approximately 30 min to more than 3 h, and varies considerably depending on the antibody and fluorophore conjugate.
- This same protocol and vacuum ring can be applied to image other organs (spleen, kidney, pancreas, skin, etc.). Ventilator is not necessary for imaging organs other than the lung.
Acknowledgments
This protocol is adapted from an earlier protocol describing live imaging of the lung (Thornton et al., 2012). This work was supported by NIH grants R01 HL118765 and R01 CA202987 (both to C.C.H.), American Heart Association Scientist Development Grant 125SDG12070005 (to R.N.H.), the La Jolla Institute for Allergy and Immunology Board of Directors Fellowship (to R.N.H.).
References
- Hanna, R. N., Cekic, C., Sag, D., Tacke, R., Thomas, G. D., Nowyhed, H., Herrley, E., Rasquinha, N., McArdle, S., Wu, R., Peluso, E., Metzger, D., Ichinose, H., Shaked, I., Chodaczek, G., Biswas, S. K. and Hedrick, C. C. (2015). Patrolling monocytes control tumor metastasis to the lung. Science 350(6263): 985-990.
- McArdle, S., Chodaczek, G., Ray, N. and Ley, K. (2015). Intravital live cell triggered imaging system reveals monocyte patrolling and macrophage migration in atherosclerotic arteries. J Biomed Opt 20(2): 26005.
- McArdle, S., Acton, S. T., Ley, K. and Ray, N. (2014). Registering sequences of in vivo microscopy images for cell tracking using dynamic programming and minimum spanning trees. IEEE International Conference on Image Processing: 3547-3551.
- Thornton, E. E., Krummel, M. F. and Looney, M. R. (2012). Live imaging of the lung. Curr Protoc Cytom Chapter 12: Unit12 28.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hanna, R. N., Chodaczek, G. and Hedrick, C. C. (2016). In vivo Imaging of Tumor and Immune Cell Interactions in the Lung. Bio-protocol 6(20): e1973. DOI: 10.21769/BioProtoc.1973.
Category
Cancer Biology > Tumor immunology > Tumor microenvironment
Immunology > Immune cell function > General
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










