- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Mouse Embryonic Fibroblast Cell Culture and Stimulation
Published: Vol 6, Iss 13, Jul 5, 2016 DOI: 10.21769/BioProtoc.1859 Views: 22233
Reviewed by: Ivan ZanoniAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification and Sorting of Adipose Inflammatory and Metabolically Activated Macrophages in Diet-Induced Obesity
Dan Wu [...] Weidong Wang
Oct 20, 2025 2232 Views

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1335 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1460 Views
Abstract
Culture of mouse embryonic fibroblast (MEF) cells represents a powerful system to test gene function due to their easy accessibility, rapid growth rates, and the possibility of a large number of experiments. Fibroblasts are a group of heterogeneous resident cells of mesenchymal origin that have various locations, diverse appearances and distinctive activities. Because of their ubiquitous distribution as tissue cells, these cells are poised to respond to factors released by newly activated innate immune cells, thus becoming a useful tool to study inflammation and immunity. Here, we describe procedures for mouse embryonic fibroblast cell isolation, primary culture, and stimulation. Specifically, we have optimized a step of serum starvation prior to stimulation. This step is necessary to maintain the quiescent status of these cells before they are exposed to pro-inflammatory stimuli for optimal responses. As shown in our previous studies, these mouse fibroblasts do not express Tnf, Csf2 or Il2 mRNAs at levels readily detectable by routine northern blotting techniques (Lai WS et al., 2006).
Materials and Reagents
- 60-mm sterile petri dish (e.g., BD Biosciences, Falcon®, catalog number: 353002 )
Note: Currently, it is “Corning, Falcon®, catalog number: 353002”. - 100-mm sterile petri dish (e.g., Corning, catalog number: 430167 )
Note: Currently, it is “Corning, Falcon®, catalog number: 430167”. - 50 ml sterile conical tube (e.g., BD Biosciences, Falcon®, catalog number: 352070 )
Note: Currently, it is “Corning, Falcon®, catalog number: 352070”. - 5 ml sterile serological pipettes (e.g., Corning Costar®, catalog number: 4051 )
- Mice (Male and female, ideally at 8-12 weeks of age)
- Ice
- Recombinant mouse tumor necrosis factor (TNF) (R&D Systems, catalog number: 410-MT )
- 1x Phosphate-buffered saline (PBS) without calcium and magnesium
- 70% ethanol
- 0.25% trypsin/EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25200-056 )
- 0.05% trypsin/EDTA (Thermo Fisher Scientific, GibcoTM, catalog number: 25300 )
- Fetal bovine serum defined (FBS) (GE Healthcare, HyClone, catalog number: SH30070.03 )
- Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 11965-092 )
- Penicillin-Streptomycin 10,000 U/ml (Thermo Fisher Scientific, GibcoTM, catalog number: 15140-122 )
- L-glutamine 200 mM (Thermo Fisher Scientific, GibcoTM, catalog number: 25030-081 )
- Rinsing medium (see Recipes)
- Complete medium (see Recipes)
- Serum-starving medium (see Recipes)
Equipment
- Dissecting microscope (e.g., Leica Microsystems, model: MZ6 )
- Dissecting instruments: tweezers and scissors
- NuAire Biological Safety Cabinet, class II, or similar
- 37 °C, 5% CO2 forced-air incubator (e.g., Thermo Fisher Scientific, FormaTM, model: 3110 )
- Centrifuge with swinging-bucket rotor and adaptors for 50-ml conical tubes
- CO2 chamber for mouse euthanization
- 37 °C shaker water bath (e.g., Thermo Fisher Scientific, PrecisionTM, model: Shaker Bath 25 )
- Autoclaved razor blades
Procedure
- Set up timed-matings between male and female mice
- Check for the plugs on the following day. The presence of a vaginal plug is considered embryonic day 0.5 (E0.5).

Figure 1. The presence of a vaginal plug is identified (arrowhead) in a female mouse on the second day following timed-mating
- Monitor weight gain in female mice every 2-3 days.
- Pregnant female mice of gestational days 14.5-15.5 are used.
Notes:- Pregnancy in female mice is usually accompanied by weight gain over time, as well as a visibly round belly.
- The weight gain for pregnant female mice at gestational day 15.5 ranges between 7.5 and 15.6 g, with an average weight gain of 11.5 g.
- Check for the plugs on the following day. The presence of a vaginal plug is considered embryonic day 0.5 (E0.5).
- Embryo removal and cell disaggregation
- Euthanize a pregnant female mouse at E14.5 or 15.5, swab abdomen with 70% ethanol, cut open the abdomen and remove the uterus, placing it in a 10-cm petri dish containing 10 ml sterile PBS on ice (Figure 2A).
- Cut away the uterine decidua, separate and transfer each embryo along with its yolk sac to a fresh 60-mm petri dish with 5 ml PBS on ice (Figure 2B-C).
- Carefully remove the yolk sac and placenta under a dissecting microscope, and examine the embryo to determine viability and/or morphological changes (Figure 2D).
Note: We routinely check for embryo size, color (pale, hemorrhage, etc.), and any obvious structural alterations including eye development and limb formation, as well as neurological abnormalities, such as spinal bifida and anencephaly. These are the two most common presentations in our experience, although both are rare in wild-type mice. - Cut the tail using clean scissors if DNA for genotyping is desired.
Note: Clean the instruments with 70% ethanol between embryos at this step to avoid carryover contamination. - Decapitate the head from the body with a pair of clean tweezers, lay the embryo on its back, and remove the visible internal organs (Figure 2D-E).
- Put the body in another sterile 60-mm petri dish containing 0.5 ml rinsing medium on ice, and move them to a tissue culture hood (Figure 2F).
- Mince the bodies with fresh, sterile razor blades into small fragments as fine as possible (Figure 2G).
- Add 5 ml of cell culture-grade Trypsin-EDTA (0.25%), and transfer to a 50 ml conical tube after pipetting up and down several times with a 5 ml pipette to disaggregate the tissue.
- Place the tube in a 37 °C water bath with shaking for enzymatic digestion for 1-2 h.
- Add another 5 ml of rinsing medium, vortex and spin at 1,200 x g for 5 min at room temperature.
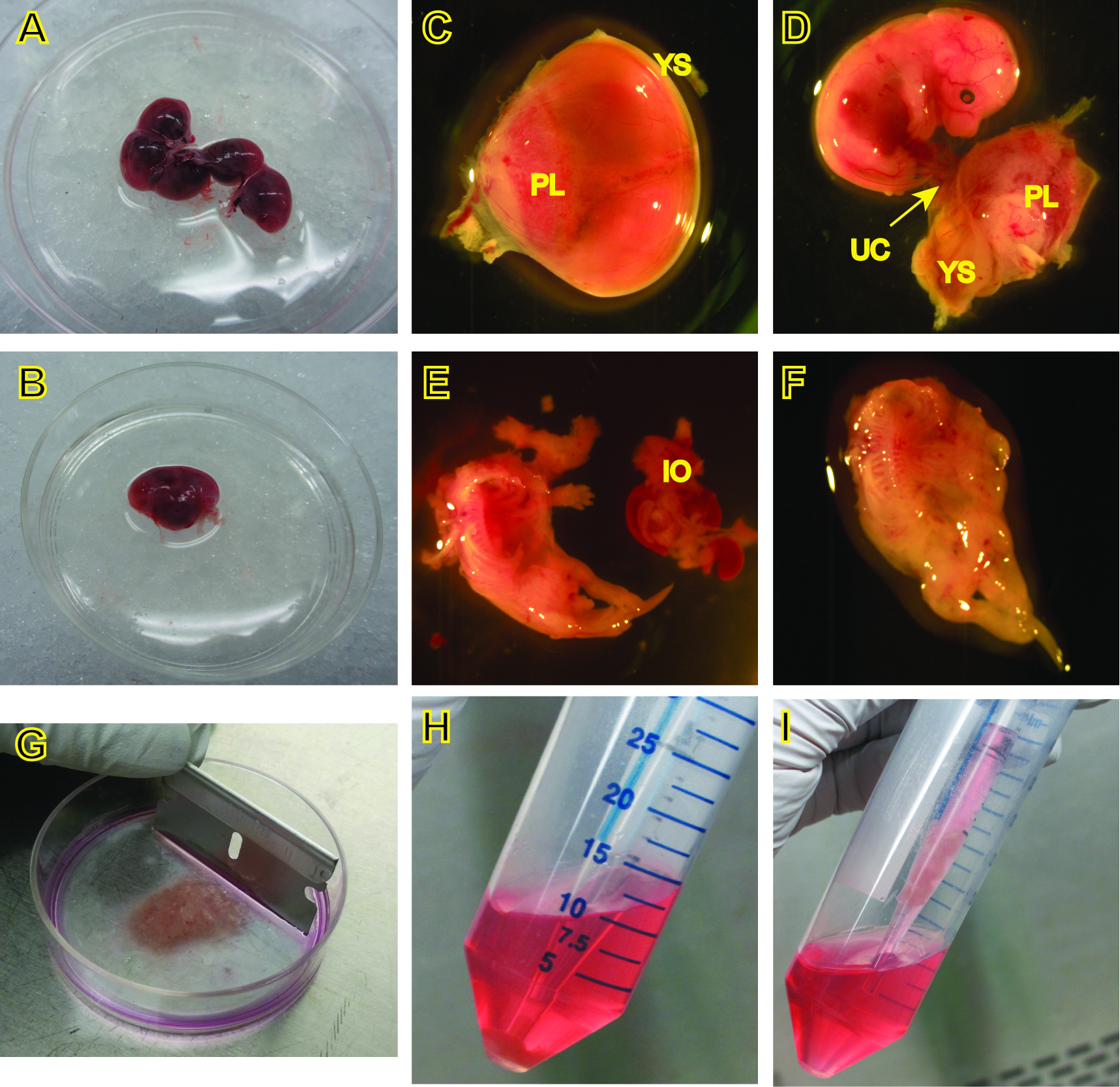
Figure 2. Procedures for the isolation of MEFs. C-F, images were captured under a dissecting microscope. PL, placenta, YS, yolk sac, UC, umbilical cord, IO, internal organs.
- Euthanize a pregnant female mouse at E14.5 or 15.5, swab abdomen with 70% ethanol, cut open the abdomen and remove the uterus, placing it in a 10-cm petri dish containing 10 ml sterile PBS on ice (Figure 2A).
- Culture and passage of mouse embryonic fibroblasts
- To obtain cells for culture, carefully place a 5 ml pipette through the supernatant and pipette up the pellet along with 0.5-1 ml of the supernatant (Figure 2H, 2I), and place in a 100-mm dish containing 10 ml complete culture medium for each embryo. Pipet cells up and down in the culture medium to disperse the cell clumps. Allow cells to adhere and grow in a 37 °C incubator supplied with 5% CO2. This is passage 0.
Note: Don’t try to aspirate the supernatant using a vacuum, since the DNA released from the cells makes it difficult to remove the supernatant without removing the cell pellets. The fibroblasts should start to attach to the bottom of dishes within 2 h after plating. - Change the medium the following day.
- Monitor cell growth. Once the cells reach confluence (~ 48 h), passage the cells at a dilution of 1:4 into 4 new 100-mm Petri dishes after washing with 10 ml of PBS, trypsinization with 2 ml of 0.05% trypsin/EDTA, and neutralization with 8 ml of complete culture medium. This is passage 1.
- Continue to expand cells when the cells are near confluence (usually within 48 h). Passage cells with trypsin treatment as above and re-plate cells into another four 100-mm Petri dishes. This is passage 2.
Note: Cells can be passaged at 1:3 to 1:5 depending on cell growth rates. The maximum number of passages for healthy primary MEFs is approximately 4, as cell growth is slowed beyond passages 4-5. We normally work with MEFs at passages 2-4 to get optimal cellular responses to stimuli.
- To obtain cells for culture, carefully place a 5 ml pipette through the supernatant and pipette up the pellet along with 0.5-1 ml of the supernatant (Figure 2H, 2I), and place in a 100-mm dish containing 10 ml complete culture medium for each embryo. Pipet cells up and down in the culture medium to disperse the cell clumps. Allow cells to adhere and grow in a 37 °C incubator supplied with 5% CO2. This is passage 0.
- Stimulation of MEFs and cell harvest
- Confluent MEFs at passages 1-3 in 100-mm Petri dishes are collected and re-plated into 60-mm Petri dishes at an initial seeding density of 2-3 x 105 cells in 5 ml culture medium per petri dish. These cells are used at passages 2-4.
Note: The estimate for the cell number in an almost confluent 100-mm petri dish is about 10-20 x 106 cells. - When the cells reach 70-80% confluence, the medium is replaced with 4.5 or 5 ml serum-starving medium for at least 16 h after one wash in serum-free DMEM.
- Add 0.5 ml serum or recombinant murine TNF into the serum-starving medium for final concentrations of 10% serum or 1-10 ng/ml of TNF as stimuli.
- Cells are then harvested for downstream applications after treatment at the indicated time points. For RNA extraction, cells are lysed with extraction buffer in dishes after one wash with ice-cold PBS. Representative data are shown in Figure 1.
- Confluent MEFs at passages 1-3 in 100-mm Petri dishes are collected and re-plated into 60-mm Petri dishes at an initial seeding density of 2-3 x 105 cells in 5 ml culture medium per petri dish. These cells are used at passages 2-4.
Representative data
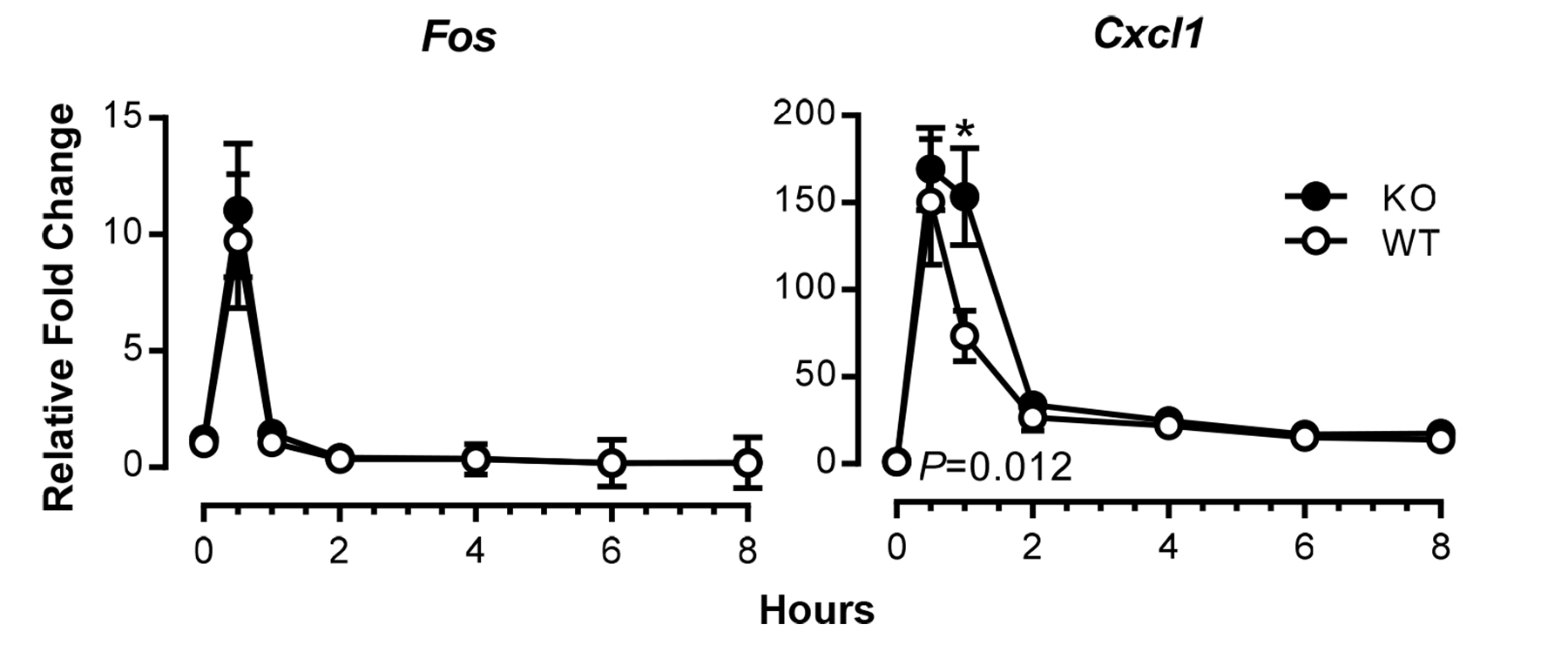
Figure 1. Time courses of induction of TNF-responsive genes in mouse fibroblasts. Expression of Fos and Cxcl1 mRNAs in serum-deprived MEFs from wild-type (WT) and tristetraprolin (TTP)-deficient (KO) mice was examined at various time points after addition of TNF at 10 ng/ml, using real-time RT-PCR. Results shown are means ± SEM of five independent experiments, and data are expressed as fold changes relative to WT at time 0; this value was set as one after normalizing to an internal control, Actb mRNA. Statistical differences between the WT and KO means, determined by paired Student t tests, are indicated (*P < 0.05, WT vs. KO). Modified from Qiu et al. (2015) with permission.
Recipes
- Rinsing medium
DMEM
1% Penicillin-Streptomycin - Complete medium
DMEM
10% FBS
1% Penicillin-Streptomycin
2 mM Glutamine - Serum-starving medium
DMEM
0.5% FBS
1% Penicillin-Streptomycin
2 mM Glutamine
Acknowledgments
This protocol was adapted from previously published studies, Lai et al. (2006) and Horner et al. (2009), and was used in Qiu et al. (2015). We thank Dee Wenzel for husbandry support, Christopher McGee for assistance with live animal imaging, and Drs. Melissa Wells and Diana Cruz-Topete for comments on the protocol. This research was supported by the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health.
References
- Horner, T. J., Lai, W. S., Stumpo, D. J. and Blackshear, P. J. (2009). Stimulation of polo-like kinase 3 mRNA decay by tristetraprolin. Mol Cell Biol 29(8): 1999-2010.
- Lai, W. S., Parker, J. S., Grissom, S. F., Stumpo, D. J. and Blackshear, P. J. (2006). Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol Cell Biol 26(24): 9196-9208.
- Qiu, L. Q., Lai, W. S., Bradbury, A., Zeldin, D. C. and Blackshear, P. J. (2015). Tristetraprolin (TTP) coordinately regulates primary and secondary cellular responses to proinflammatory stimuli. J Leukoc Biol 97(4): 723-736.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Qiu, L. Q., Lai, W. S., Stumpo, D. J. and Blackshear, P. J. (2016). Mouse Embryonic Fibroblast Cell Culture and Stimulation. Bio-protocol 6(13): e1859. DOI: 10.21769/BioProtoc.1859.
Category
Immunology > Immune cell isolation > Maintenance and differentiation
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









