- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparation of Knockdown Transformants of Unicellular Charophycean Alga, Closterium peracerosum-strigosum-littorale Complex
Published: Vol 6, Iss 10, May 20, 2016 DOI: 10.21769/BioProtoc.1813 Views: 9784
Reviewed by: Maria SinetovaChristian SailerAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

PEG-mediated, Stable, Nuclear and Chloroplast Transformation of Cyanidioschizon merolae
Maksymilian Zienkiewicz [...] Kinga Kania
Sep 5, 2019 4967 Views

A Fast and Easy Method to Study Ralstonia solanacearum Virulence upon Transient Gene Expression or Gene Silencing in Nicotiana benthamiana Leaves
Wenjia Yu and Alberto P. Macho
Aug 5, 2021 4699 Views

Agrobacterium-mediated Genetic Transformation of Cotton and Regeneration via Somatic Embryogenesis
Alka Srivastava [...] Praveen C. Verma
May 20, 2023 4333 Views
Abstract
To prepare the knockdown transformants of the Closterium peracerosum-strigosum-littorale (C. psl.) complex, particle bombardment was applied with a newly constructed vector (pSA0104) with an endogenous constitutive promoter fused to a DNA fragment corresponding to an antisense strand of a target gene. Using a hygromycin resistance gene (aph7”), hygromycin-resistant colonies were selected. After the second screening, integration of the vector into the genome was checked by PCR and the knockdown effect was evaluated by Western blotting using a specific antibody against the target protein.
Keywords: KnockdownMaterials and Reagents
- 0.6 µm gold microcarriers (Bio-Rad Laboratories, catalog number: 165-2262 ) or 0.25 µm gold nanoparticle (BBI Solutions, catalog number: EMGC250 )
- Cell culture dishes, 90 mm x 20 mm polystyrene (AGC TECHNO GLASS CO., catalog number: SH90-20 )
- 50 ml culture tube (Thomas Scientific, Labcon, catalog number: 3181-345-008 )
- 1.5 ml microtube (BMbio, catalog number: BM-15 )
- Parafilm (VWR International, Bemis, catalog number: PM996 )
- Micropore surgical tape (3M, catalog number: 1530-0 )
- Qualitative filter paper No. 2 (Toyo Roshi Kaisha, Advantec, catalog number: 00021110 )
- Test tube for incubation of transformant (AGC TECHNO GLASS CO., catalog number: TST-SCR16-150 )
- pSA0104 Vector
- Species: Heterothallic C. psl. complex strains [NIES-67 (mt+) and NIES-68 (mt-) (the National Institute for Environmental Studies)]
- KOD-plus NEO DNA polymerase (TOYOBO CO., catalog number: KOD-401 )
- KOD-FX DNA polymerase (TOYOBO CO., catalog number: KFX-101 )
- GENEART seamless cloning and assembly kit (Thermo Fisher Scientific, InvitrogenTM, catalog number: A13288 )
- High Pure Plasmid Isolation kit (Roche Diagnostics, catalog number: 11754785001 )
- Spermidine (Wako Pure Chemical Industries, catalog number: 191-13831 )
- Agar powder (Nacalai tesque, catalog number: 01028-85 )
- Ethanol absolute (Wako Pure Chemical Industries, catalog number: 057-00451 )
- Glycerol (Wako Pure Chemical Industries, catalog number: 075-00616 )
- Quant-iT dsDNA Assay Kit, broad range (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q-33130 )
- Hygromycin B (Wako Pure Chemical Industries, catalog number: 085-06153 )
- QuickExtract Plant DNA Extraction Solution (Epicentre, catalog number: QEP80705 )
- Calcium nitrate tetrahydrate [Ca(NO3)2.4H2O] (Wako Pure Chemical Industries, catalog number: 039-00735 )
- Potassium nitrate (KNO3) (Wako Pure Chemical Industries, catalog number: 160-04035 )
- Disodium β-glycerophosphate pentahydrate (Sigma-Aldrich, catalog number: 50020-1000 G )
- Magnesium sulfate heptahydrate (MgSO4.7H2O) (Nacalai tesque, catalog number: 21003-75 )
- Vitamin B12 (Wako Pure Chemical Industries, catalog number: 226-00343 )
- Biotin (Wako Pure Chemical Industries, catalog number: 023-08711 )
- Thiamine HCl (Wako Pure Chemical Industries, catalog number: 201-00852 )
- 2-amino-2-hydroxymethyl-1, 3-propanediol (Wako Pure Chemical Industries, catalog number: 011-16381 )
- Hydrochloric acid (HCl) (Nacalai tesque, catalog number: 18321-05 )
- Na2EDTA.2H2O (Wako Pure Chemical Industries, catalog number: 345-01865 )
- Iron(III) chloride hexahydrate (FeCl3.6H2O) (Wako Pure Chemical Industries, catalog number: 091-00872 )
- Manganese(II) chloride tetrahydrate (MnCl2.4H2O) (Nacalai tesque, catalog number: 21211-45 )
- Zinc sulfate heptahydrate (ZnSO4.7H2O) (Nacalai tesque, catalog number: 37011-62 )
- CoCl2.6H2O (Wako Pure Chemical Industries, catalog number: 003-00368 )
- Na2MoO4.2H2O (Wako Pure Chemical Industries, catalog number: 019-00247 )
- Calcium chloride dihydrate (CaCl2.2H2O) (Wako Pure Chemical Industries, catalog number: 039-00431 )
- C medium (see Recipes)
- PIV metals (see Recipes)
- Conditioned C medium (see Recipes)
- MI medium (see Recipes)
Equipment
- 300 ml Erlenmeyer flask (AGC TECHNO GLASS CO., catalog number: 4980FK300 )
- Growth chamber (Nippon Medical & Chemical Instruments, model: KCLP-1400II CT ), being discontinued
- Handmade hemocytometer (1 x 1 mm, grid length x grid width) (not commercially available)
- Thermal cycler (Thermo Fisher Scientific, Applied BiosystemsTM, model: veriti200)
- Centrifuge (Hitachi Ltd., model: CF16RX )
- Swing rotor (Hitachi Ltd., model: T5SS31 )
- Angle rotor (Hitachi Ltd., model: T15AP31 )
- Centrifuge (KUBOTA Corporation, model: 1920 ), being discontinued
- Angle rotor (KUBOTA Corporation, model: RA-48J ), being discontinued
- Cute mixer (EYELA, model: CM-1000 )
- Qubit fluorometer (Thermo Fisher Scientific, InvitrogenTM, catalog number: Q32857 ), being discontinued
- Ultrasonic cleaner (Sigma-Aldrich, Branson®, model: 3510J-DTH )
- Gene transfer system (Tanaka co., model: IDERA GIE-III )
- Fluorescence stereomicroscope LEICA MZ16 F (Leica Microsystems)
- Micro Pick and Place System (Nepa Gene Co., catalog number: MPP-300 )
- Light microscope (Olympus, model: CK-40 ), being discontinued
Procedure
- Preparation of cells for transformation
- Culture vegetative cells in 150 ml C medium (http://www.nies.go.jp/biology/mcc/home.htm) at 23 °C under a 16 h light (28 µmol/m2/s) and 8 h dark cycle in 300 ml Erlenmeyer flasks.
- Collect the C. psl. complex cells from the mid-logarithmic to early stationary phase (9-11 days of culture) in a 50 ml culture tube and concentrate by centrifugation (1,100 x g for 2 min at 23 °C) using a swing-type rotor (T5SS31). For the slowdown, deceleration should be set to the slowest speed.
- Re-suspend the cells in a small aliquot of fresh C medium and count the cells under a light microscope (CK40) using a handmade hemocytometer. Adjust to a density of 5 x 106 cells ml-1 by adding fresh C medium.
- Spread the cell suspension (1 x 106 cells) using a glass bacterial spreader onto a 90-mm plate containing C medium with 1.5% agar (w/v), prepared one day previously and stored at room temperature.
- Culture the cells at 23 °C for 2 days under continuous light at 10 µmol m-2 s-1 (Figure 1).
Figure 1. Photograph of gold particles sedimented by centrifugation
- Culture vegetative cells in 150 ml C medium (http://www.nies.go.jp/biology/mcc/home.htm) at 23 °C under a 16 h light (28 µmol/m2/s) and 8 h dark cycle in 300 ml Erlenmeyer flasks.
- Preparation of constructs for transformation of the C. psl. complex (approximately 4-5 days)
- To avoid off-target silencing, the DNA region used for the antisense expression should be carefully checked, using the BLASTN program, against unpublished RNAseq and a genome database of the C. psl. complex.
- Amplify the required region of target DNA by PCR with KOD-plus-NEO DNA polymerase from plasmid clones (2 min at 94 °C, followed by 30 cycles of 10 sec at 98 °C and 1 min/kbp at 68 °C). In the case of CpRLK1 gene, 2,180 bp fragment, encoding the extracellular domain of CpRLK1 protein, was amplified. The primer DNAs should include extra sequences corresponding to the vector sequences (ex. 5'-ccagcatgactagtctcgagTTCGGGCTGTTGCTTCGGCGTCA-3' and 5'-gcttcatcaaattactcgagTGGGTGCCGCCGTAGGTTAATAT-3'), which are required for cloning into the pSA0104 vector (Figure 2, Hirano et al., 2015) using the GENEART kit. The enzyme mix provided with the kit recognizes and assembles the vector and PCR fragments sharing terminal end-homology. The pSA0104 vector contains a promoter region of CpHSP70 (5’CpHSP70) for expression of the target gene and a hygromycin resistance gene (aph7”, Berthold et al., 2002) for selection (Figure 2).
A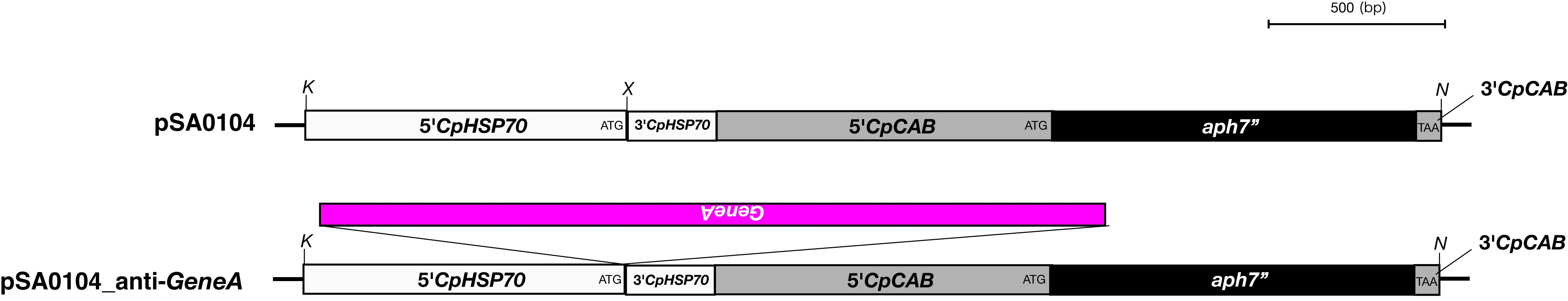
B
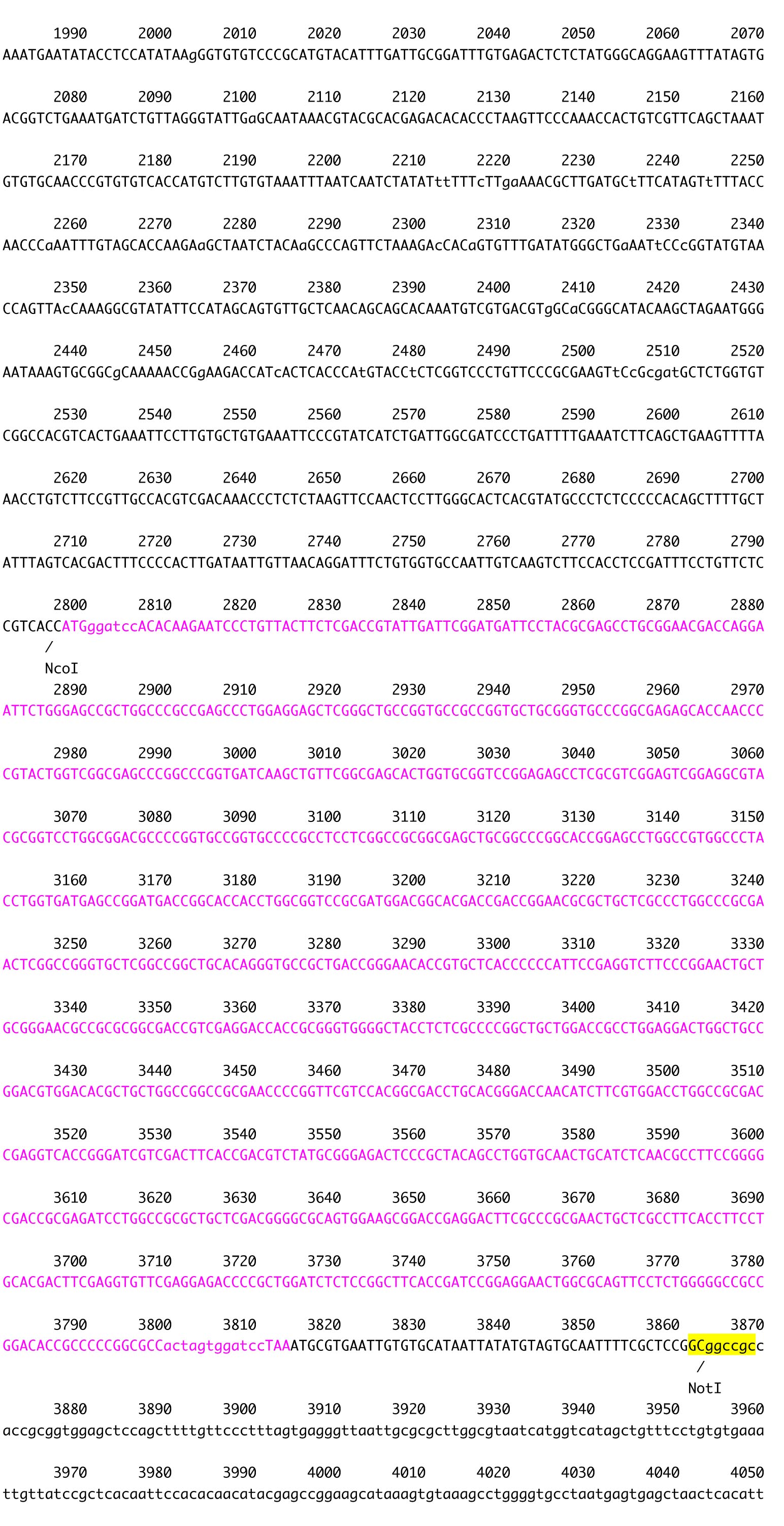
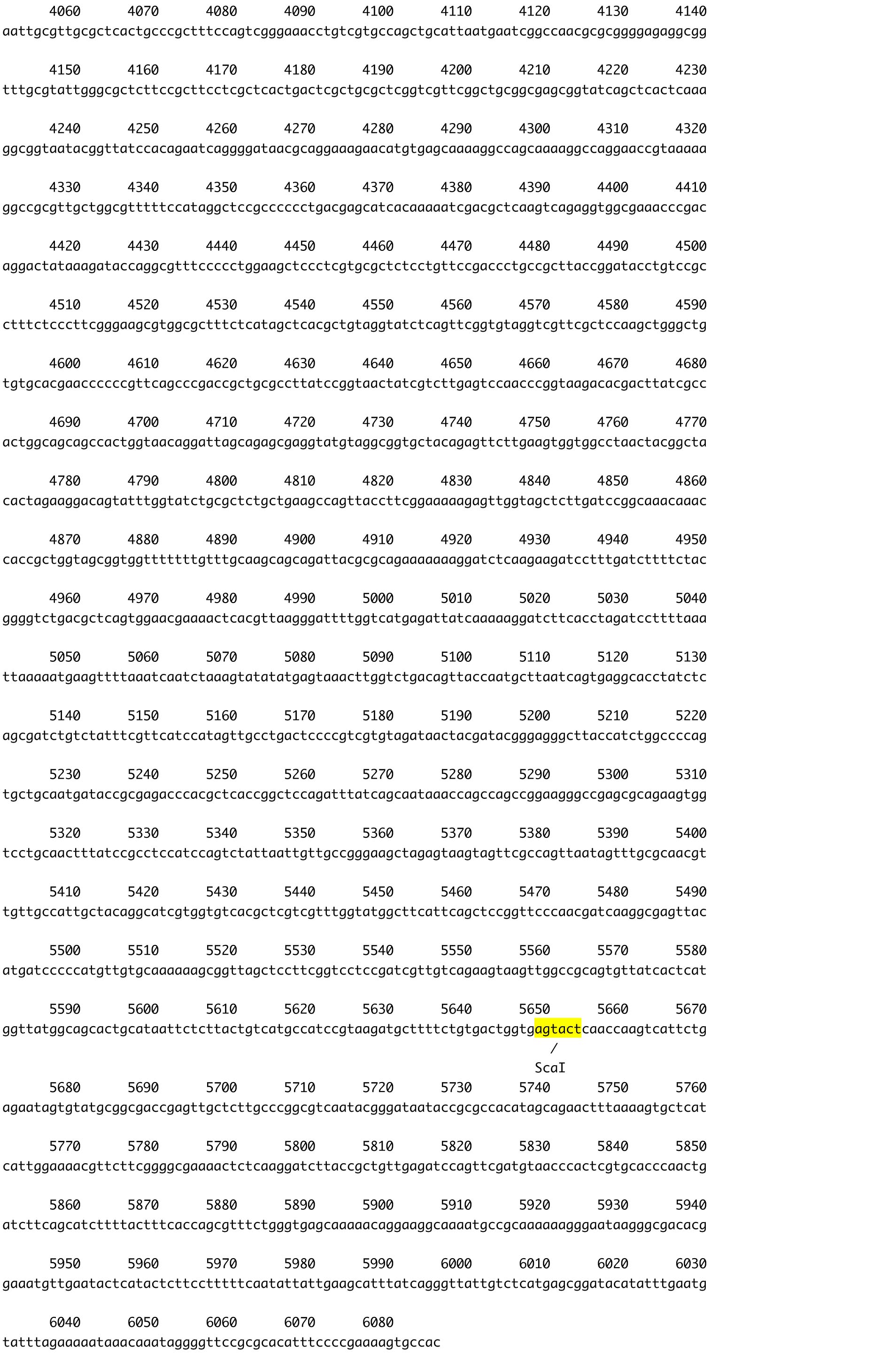
Figure 2. The plasmid vector for transformation of C. psl. complex. A. Constructs of pSA0104 and pSA0104_anti-GeneA. The promoters and untranslated regions of CpCAB1 and CpHSP70 genes are shown as light gray and white boxes, respectively. The aph7’’ gene (hygromycin resistance gene) is indicated by black box. Forward and reverse primers used for plasmid construction are shown as arrows under the boxes. Initiation codon (ATG) and stop codon (TAA) are indicated in their respective boxes. The plasmid backbone of the construct was pBluescript II SK+. K, KpnI; N, NotI; X, XhoI. B. The sequence of pSA0104. The restriction sites used for cloning and for linearization have been highlighted. Blue character indicates the promoter and untranslated regions of CpCAB1. Pink character indicates aph7’’ gene. - Using the GENEART kit, insert the amplified DNA fragment corresponding to the antisense strand of a target gene into the XhoI site (immediately after pCpHSP70) of the pSA0104 vector.
- After confirmation of the sequence, prepare a sufficient amount of plasmid DNA from an E. coli culture using a High Pure Plasmid Isolation kit, or the standard Alkaline SDS method, followed by phenol/chloroform purification (Birnboim and Doly, 1979).
- Quantify the DNA concentration accurately using a Qubit fluorometer and the Quant-iT dsDNA Assay Kit.
- To linearize the constructs, digest the vector arm region using suitable restriction enzymes, according to the manufacturer’s instruction. In most cases, NotI, KpnI, or ScaI (on pBluescript II SK+) can be used (Figure 2).
- To avoid off-target silencing, the DNA region used for the antisense expression should be carefully checked, using the BLASTN program, against unpublished RNAseq and a genome database of the C. psl. complex.
- Microcarrier stock preparation
- Add 60 mg of gold particles and 1 ml of 70% (w/v) ethanol solution to a 1.5 ml microtube.
Note: 0.25 µm of gold particles would result in a higher transformation efficacy than 0.6 µm (Abe et al., 2008), however, this may be expensive. Normally, 0.6 µm gold particles are used. - Vortex at 2,000 rpm using a micromixer for 5 min.
- Allow to sediment for 15 min at room temperature.
- Further sediment by centrifugation using a T15AP31 rotor at 18,800 x g for 10 secfI.
- Carefully remove the solution with a pipette.
- Add 1 ml of sterile MilliQ water and vortex for 2 min. Sediment by centrifugation as in step B4. Discard the water. Repeat 3 times.
- Add 1 ml of 50% sterile glycerol solution and vortex for 1 min.
- Stored at -20 °C until needed.
- Add 60 mg of gold particles and 1 ml of 70% (w/v) ethanol solution to a 1.5 ml microtube.
- Coating the microcarriers with the plasmid DNA
- Thaw the microcarriers (step C8) and suspend by sonication for 5 min using an ultrasonic cleaner (set the mode “sonics”).
- Add the following reagents to the 82.5 µl (4.95 mg, for 20 shots) of microcarrier suspension in a 1.5 ml microtube. Mix by vortexing briefly after the addition of each reagent.
- 82.5 µl of linearized construct (for 400 ng/µl of empty pSA0104, which is 6,086 bp in length). Adjust the concentration to the equal mol value depending on the size of the construct.
- 250 µl of 2.5 M CaCl2 (sterile filtered)
- 100 µl of 0.1 M spermidine (sterile filtered)
- 82.5 µl of linearized construct (for 400 ng/µl of empty pSA0104, which is 6,086 bp in length). Adjust the concentration to the equal mol value depending on the size of the construct.
- Vortex for 2 min.
- Incubate for 30 min at room temperature. Invert the tube gently every 10 min.
- Sediment by centrifugation at 8,300 x g for 10 sec (RA-48J rotor) using a Kubota 1920 centrifuge.
- Discard the supernatant without disturbing the microcarrier sediment.
- Add 750 µl of 70% (v/v) ethanol and vortex for 2 min.
- Sediment and discard the supernatant as in steps D5-6.
- Add 750 µl of absolute ethanol and vortex for 2 min.
- Sediment and discard the supernatant as in steps D5-6.
- Add 250 µl of absolute ethanol. Seal the cap with parafilm to minimize ethanol evaporation until needed.
- Thaw the microcarriers (step C8) and suspend by sonication for 5 min using an ultrasonic cleaner (set the mode “sonics”).
- Particle bombardment
Note: This protocol is optimized for the IDERA GIE-III gene transfer system (Figure 3). Further optimization may be required if other particle delivery systems are used.
Figure 3. Photograph of IDERA GIE-III gene transfer system- Prepare an autoclaved top agar solution [0.4% (w/v) agar in C medium] and incubate the melted agar solution in a water bath at 42 °C.
- Place a barotolerant chamber on a clean bench (Figure 4).
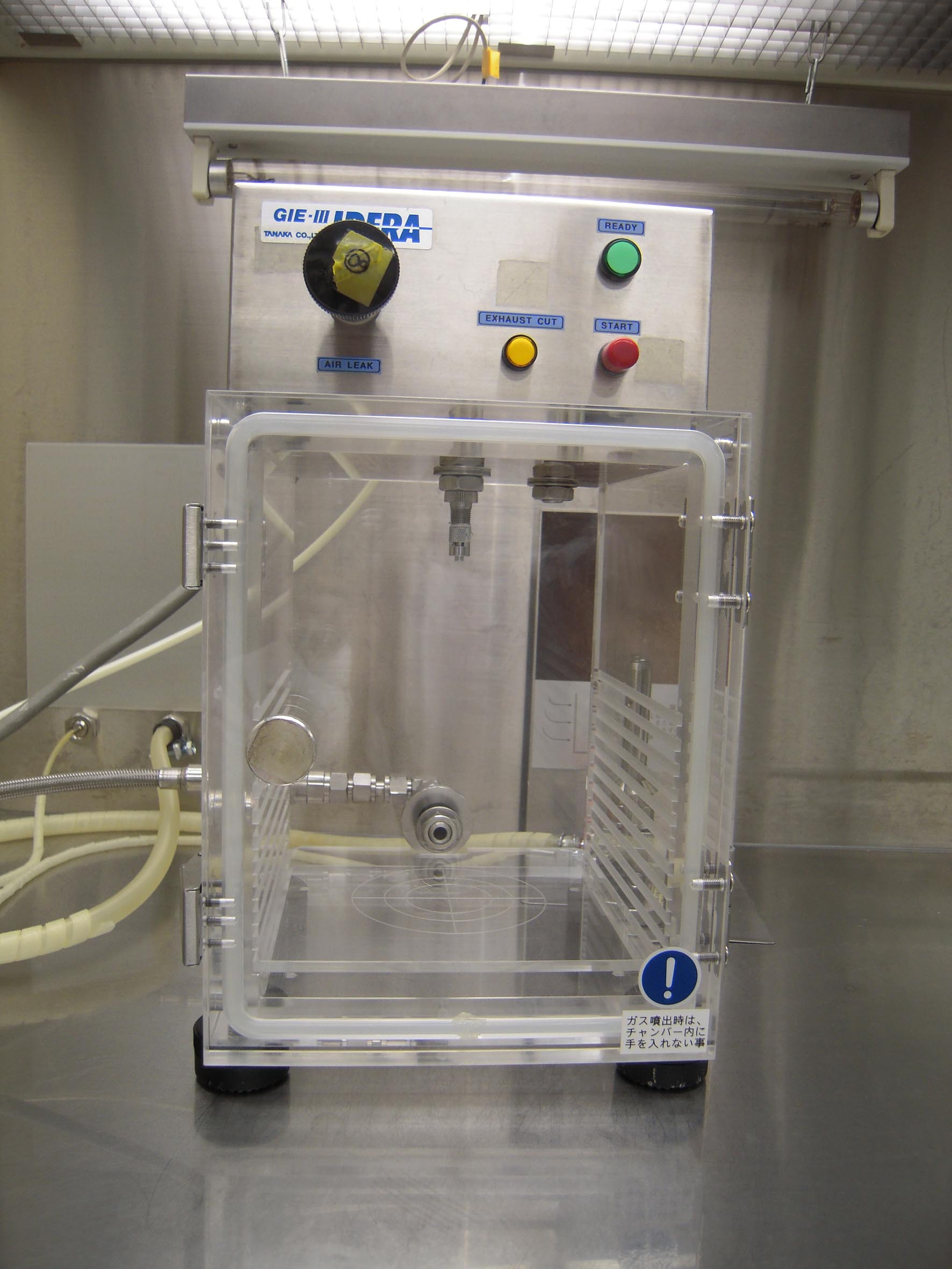
Figure 4. Photograph of barotolerant chamber on a clean bench - Suspend the microcarriers (step D11) completely by vortexing.
- Add 3.3 µl of microcarrier suspension into each of three wells (Figure 5) of autoclaved DNA cartridges.
Note: Before adding the suspension, we recommend bombarding the empty cartridges to empty the dishes. This step cleans and prevents clogging of the wells.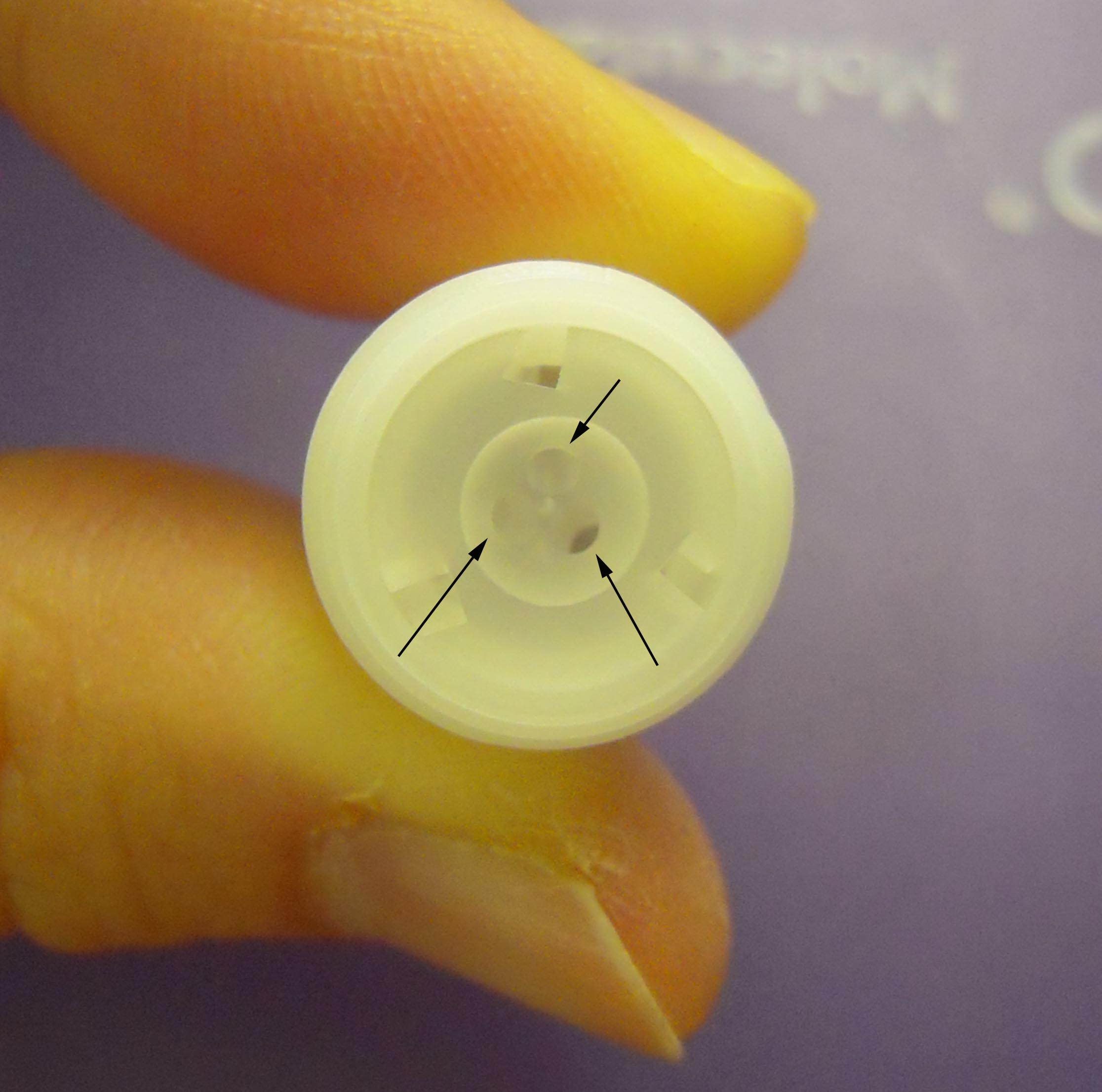
Figure 5. Photograph of DNA cartridge focusing on three wells. The arrows indicate the wells. - Place the DNA cartridge and precultured C. psl. complex on the culture dish (step A5, Figure 6) into the chamber (Figure 7).

Figure 6. Photograph of Closterium cells cultured for 2 days on 90-mm plate containing C medium with 1.5% agar (w/v)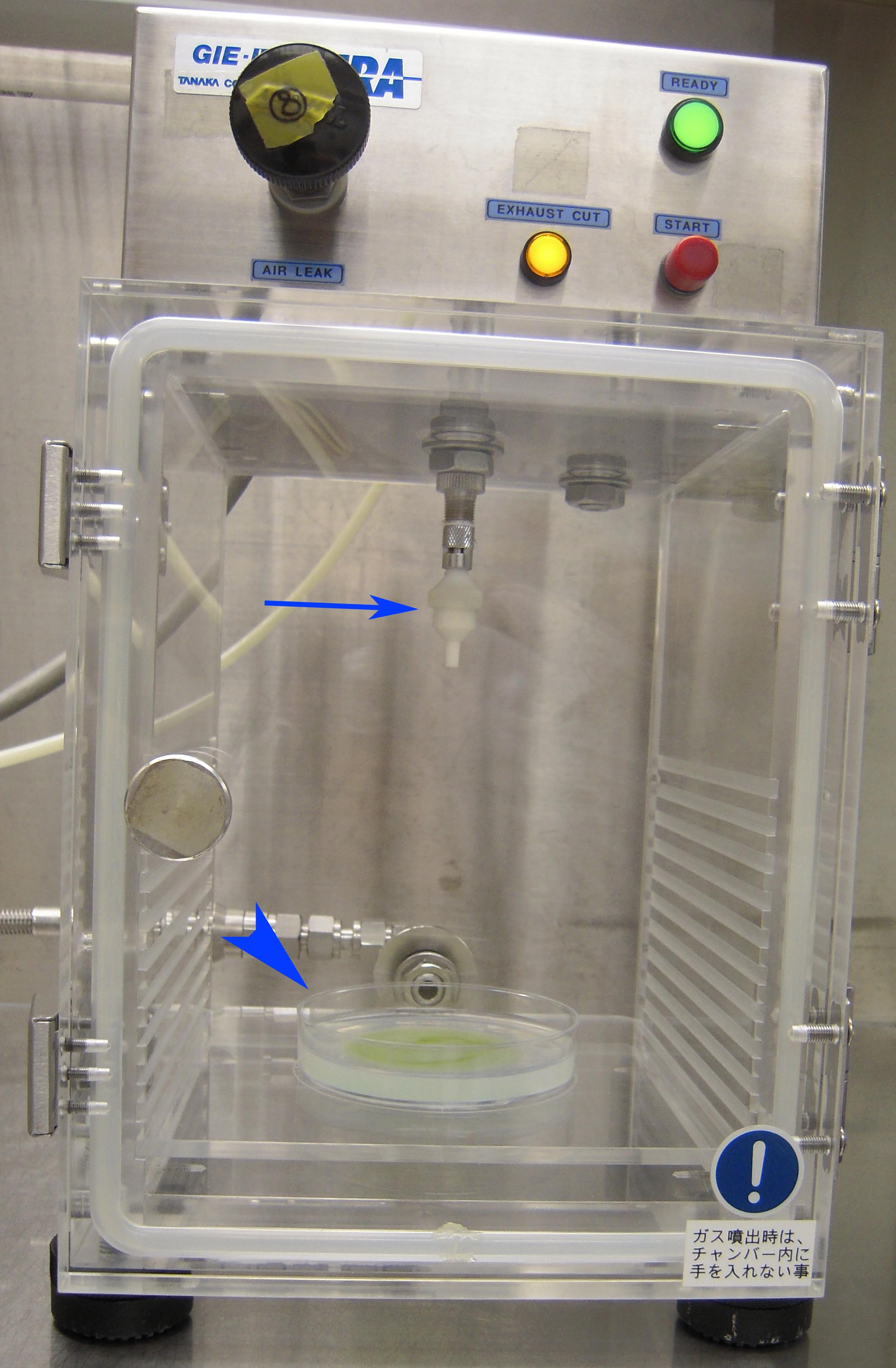
Figure 7. Photograph of barotoletant chamber just before the bombardment. Arrow indicates the DNA cartridge and arrowhead indicates the cultured plate. - Start bombardment using the following parameters (Figure 8):
Gun-to-target distance 13.0 cm Helium exit pressure 5.5 kgf/cm Vacuum in the barotolerant chamber 710-715 mmHg Opening time of gas valve to allow rapid flow of helium gas 0.05 sec 
Figure 8. Photograph of IDERA GIE-III focusing on the parameters
- Prepare an autoclaved top agar solution [0.4% (w/v) agar in C medium] and incubate the melted agar solution in a water bath at 42 °C.
- Selection of hygromycin resistant cells
- Immediately after bombardment, add 5 ml of melted top agar (step E1) and allow to stand for 30 min.
- Add 4 ml of C medium to the agar plate. Seal the plate using surgical tape and incubate for 2 d at 23 °C under continuous light (10 µmol m-2 s-1) to enable accumulation of the aminoglycoside phosphotransferase (aph’’ protein).
- Carefully discard the C medium on the agar by decantation. Do not disturb the top surface of the agar.
- Add 5 ml of C medium containing 50-100 µg ml -1 hygromycin B to the plate to select the hygromycin-resistant colonies.
Note: The suitable concentration of hygromycin B for screening should be tested before transformation as this depends on the strains used. In our experience, a lower concentration (10 µg ml-1) is suitable to select for certain C. psl. complex strains. - Incubate the cells for 3 to 5 weeks, with a weekly replacement of fresh. hygromycin-containing medium, under continuous light at 23 °C.
- Pick the surviving colonies (Figure 9) and streak onto 0.8% (w/v) agar plates containing conditioned C medium (Abe et al., 2011) and hygromycin. The unknown factor(s) for cell proliferation, which was secreted from growing cells into surrounded environment, would be included in the conditioned medium and would facilitate the cell division.
Note: Use a stereomicroscope to check for contamination of the colonies with bacteria and/or fungi (Figure 10). If contamination occurs, wash the colony repeatedly (at least three times) by the capillary washing method (Andersen and Kawachi, 2005) or isolate a single cell using the Micro Pick and Place System.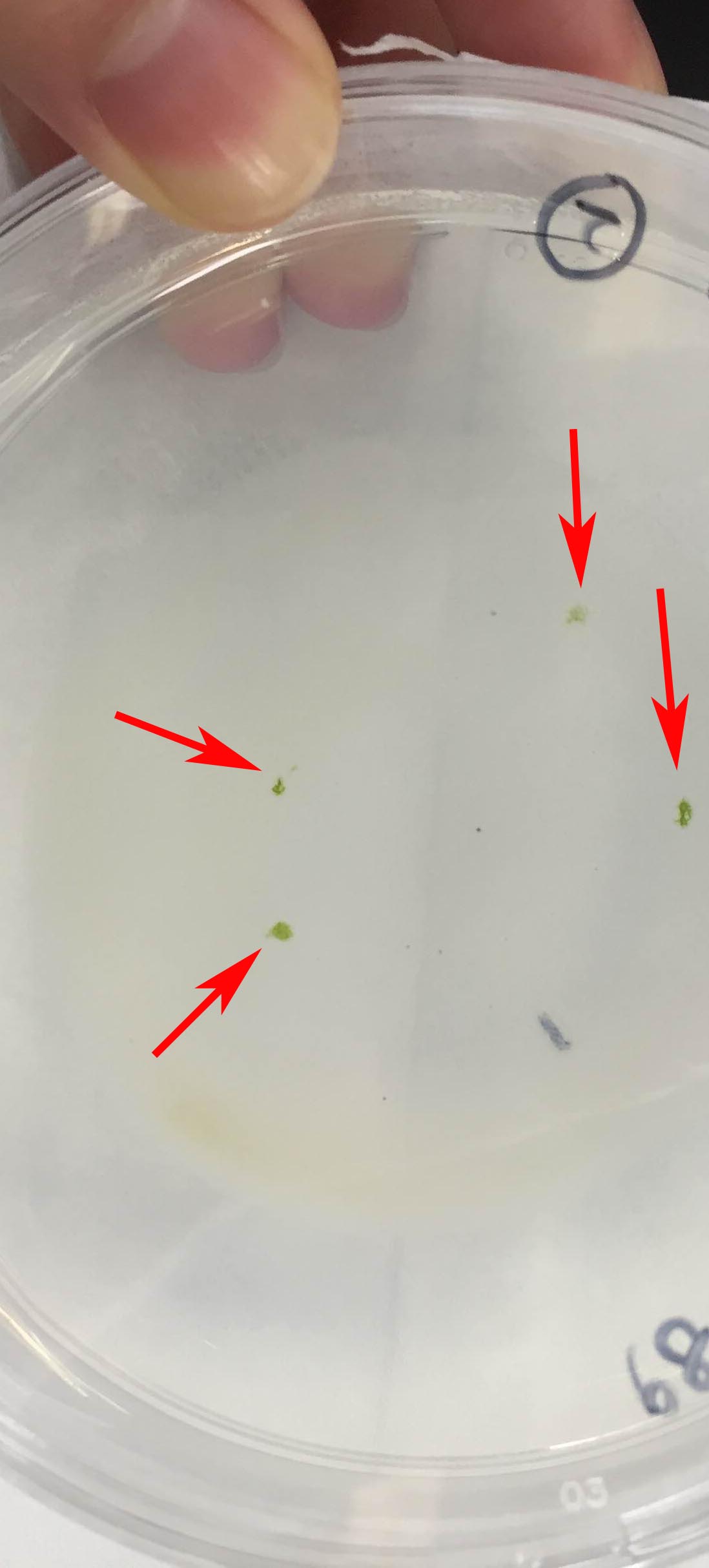
Figure 9. Photograph of surviving colonies after the selection. Arrows indicate the survived colonies, which have been incubated in C medium containing 50 µg ml-1 hygromycin B for 5 weeks.
Figure 10. Photograph of a contaminated colony. Scale bar = 200 µM - Pick the single colony on the plates (step F6) and transfer to 5 ml of conditioned medium in a test tube. Incubate at 23 °C under a 16 h light and 8 h dark cycle for 2-3 weeks.
Note: If contamination occurs, washing and isolation of a single cell is required as described in Note of step F6. - Transfer the proliferated cells from step F7 to fresh C medium and maintain under normal conditions (step A1).
- Immediately after bombardment, add 5 ml of melted top agar (step E1) and allow to stand for 30 min.
- Confirmation of successful integration of the constructs into the genome
- Isolate crude genomic DNA from cells using QuickExtract Plant Extraction Solution, according to the manufacturer’s instruction.
- Amplify the integrated DNA from the genomic DNA by PCR with KOD FX DNA polymerase (2 min at 94 °C, followed by 40 cycles of 15 sec at 98 °C and 1 min/kbp at 68 °C).
- Isolate crude genomic DNA from cells using QuickExtract Plant Extraction Solution, according to the manufacturer’s instruction.
- Evaluation of knockdown effects
- If specific antibody against the target protein is being used, check the protein expression levels by Western blotting.
- Select those transformants showing a high reduction in protein expression levels for further characterization, because the knockdown effect will vary and some will not show a distinct reduction in protein expression.
- Evaluate the phenotype of the selected knockdown transformants. In our case, the sexual responses of transformants in MI medium was mostly evaluated.
- If specific antibody against the target protein is being used, check the protein expression levels by Western blotting.
Recipes
- C medium
Stock solution for 5 L of C media
Adjust pH to 7.5 with HCl10% (w/v) Ca(NO3)2.4H2O solution 7.5 ml 5% (w/v) KNO3 solution 10 ml 5% (w/v) disodium β-glycerophosphate pentahydrate solution 5 ml 4% (w/v) MgSO4.7H2O solution 5 ml 0.0001% (w/v) vitamin B12 solution 0.5 ml 0.0001% (w/v) biotin solution 0.5 ml 0.0112% (w/v) thiamine HCl solution 0.45 ml PIV metals 15 ml 2-amino-2-hydroxymethyl-1, 3-propanediol 2.5 g
a.PIV metals
Component (for 500 ml)Na2EDTA.2H2O 500 mg FeCl3.6H2O 98 mg MnCl2.4H2O 18 mg ZnSO4.7H2O 11 mg CoCl2.6H2O 2 mg Na2MoO4.2H2O 1.25 mg - Conditioned C medium (Abe et al., 2011)
Incubate wild-type mt+ or mt- cells in fresh C medium for 14-20 days
Collect the cultured medium by filtration using qualitative filter paper and sterilize the filtered medium by autoclaving (121 °C for 15 min) - MI medium
Stock solution for 5 L of MI media
Adjust pH to 8.5 with HCl5% (w/v) CaCl2.2H2O solution 10 ml 5% (w/v) disodium β-glycerophosphate pentahydrate solution 5 ml 4% (w/v) MgSO4.7H2O solution 5 ml 0.0001% (w/v) vitamin B12 solution 0.5 ml 0.0001% (w/v) biotin solution 0.5 ml 0.0112% (w/v) thiamine HCl solution 0.45 ml PIV metals 15 ml 2-amino-2-hydroxymethyl-1, 3-propanediol 2.5 g
Acknowledgments
The authors wish to thank Dr. Kensuke Ichihara (Univ. Tokyo) and Ms. Wakana Takiguchi (JWU) for their technical supports. This work was partly supported by Grants-in-Aid for Scientific Research (nos. 24370038, 24247042, 25304012, 26650147, 15H05237 to H.S., no. 23770277 to J. A., nos. 23770093 and 26440223 to Y. T.) from the Japan Society for the Promotion of Science, Japan, a grant from the New Technology Development Foundation to H. S. and Y. T., and MEXT-supported Program for the Strategic Research Foundation at Private Universities to H. S.
References
- Abe, J., Hiwatashi, Y., Ito, M., Hasebe, M. and Sekimoto, H. (2008). Expression of exogenous genes under the control of endogenous HSP70 and CAB promoters in the Closterium peracerosum-strigosum-littorale complex. Plant Cell Physiol 49(4): 625-632.
- Abe, J., Hori, S., Tsuchikane, Y., Kitao, N., Kato, M. and Sekimoto, H. (2011). Stable nuclear transformation of the Closterium peracerosum-strigosum-littorale complex. Plant Cell Physiol 52(9): 1676-1685.
- Andersen, R. A. and Kawachi, M. (2005) Chapter 6: Traditional microalgae isolation techniques, In: Andersen R. A. (ed). Algal Culturing techniques. Elsevier Academic Press, pp 83-100 (total 578 pages).
- Berthold, P., Schmitt, R. and Mages, W. (2002). An engineered Streptomyces hygroscopicus aph 7" gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153(4): 401-412.
- Birnboim, H. C. and Doly, J. (1979). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7(6): 1513-1523.
- Hirano, N., Marukawa, Y., Abe, J., Hashiba, S., Ichikawa, M., Tanabe, Y., Ito, M., Nishii, I., Tsuchikane, Y. and Sekimoto, H. (2015). A receptor-like kinase, related to cell wall sensor of higher plants, is required for sexual reproduction in the unicellular charophycean alga, Closterium peracerosum-strigosum-littorale complex. Plant Cell Physiol 56(7): 1456-1462.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Abe, J., Hirano, N., Komiya, A., Kanda, N., Fujiwara, A., Hori, S., Tsuchikane, Y. and Sekimoto, H. (2016). Preparation of Knockdown Transformants of Unicellular Charophycean Alga, Closterium peracerosum-strigosum-littorale Complex. Bio-protocol 6(10): e1813. DOI: 10.21769/BioProtoc.1813.
Category
Molecular Biology > DNA > Transformation
Plant Science > Phycology > DNA > Transformation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link