- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Identification of RNA-binding Proteins by RNA Ligand-based cDNA Expression Library Screening
Published: Vol 6, Iss 2, Jan 20, 2016 DOI: 10.21769/BioProtoc.1715 Views: 8696
Reviewed by: Antoine de MorreeZhen Shi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
Saurabh Upadhyay
Aug 20, 2025 2441 Views

Isolation of Antigen-Specific Nanobodies From Synthetic Libraries Using a Protein Selection Strategy That Combines MACS-Based Screening of YSD and FLI-TRAP
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
Jan 20, 2026 443 Views
Abstract
We previously reported when a portion of the Requiem (REQ/DPF2) messenger ribonucleic acid (mRNA) 3’ untranslated region (3’UTR), referred to as G8, was overexpressed in K562 cells, β-globin expression was induced, suggesting that the 3’UTR of REQ mRNA plays a physiological role (Kim et al., 2014). To identify trans-acting factors that bind to the REQ 3’UTR, we describe the RNA ligand based cDNA expression library screening method. This protocol could be adapted to detect specific RNA-protein interactions. Following this method, we identified six positive clones in the initial round of screening and four pure clones after sib-screening. This protocol was originally published in Kim et al. (2014).
Keywords: RNA-protein interactionMaterials and Reagents
- X-Omat AR film (Eastman Kodak Company, catalog number: 0572842 )
- Eppendorf tubes
- K562 cells
Note: Phagemid-based K562 cDNA expression libraries were constructed by isolating mRNA from cells with an Ultraspec-RNA isolating system and a biotinylated oligo (dT) probe. - XL1-Blue cells (Agilent Technologies, catalog number: 200403 )
- XLOR cell (Agilent Technologies, catalog number: 200403 )
- 10% bovine calf serum (GE Healthcare, HycloneTM, catalog number: SH30073.03 )
- RPMI 1640 medium (Life Technologies, GibcoTM, catalog number: 11875-093 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 11875-093”. - Ultraspec-RNA isolating system (Biotech, catalog number: BL-10050 )
- Biotinylated oligo (dT) probe (50 pmol/μl) (Promega Corporation, catalog number: Z5261 )
- cDNA synthesis kit (Agilent Technologies, catalog number: 200403 )
- λZAP II express phage vector (Agilent Technologies, catalog number: 200403 )
- Nitrocellulose membrane (immobilon-NC membrane) (EMD Millipore Corporation, catalog number: N8395 )
- Ribonucleic acid from torula yeast (RNA type VI) (Sigma-Aldrich, catalog number: R6625 )
- [α-32P]-labeled G8-RNA ligand (BMS)
- ExAssist helper phage (Agilent Technologies, catalog number: 200253 )
- IPTG [≥ 99% (TLC), ≤ 0.1% Dioxan] (Sigma-Aldrich, catalog number: I6758 )
- HEPES (pH 7.9)
- KCl
- 0.1% (w/v) Ficoll 400-DL
- 0.01% polyvinyl-pyrolidon PVP-40
- MnCl2
- ZnCl2
- EDTA
- DTT
- Tryptone
- NaCl
- Yeast Extract
- DW
- MgSO4
- Tris-HCl (pH 7.5)
- Gelatin
- Screening buffer (see Recipes)
- LB plates (see Recipes)
- SM buffer (see Recipes)
Equipment
- Refrigerated Eppendorf centrifuge (Hanil, catalog number: Union 55R )
- Heat block (Thermolyne, catalog number: DB17615 )
- Tissue culture CO2 incubators set at 37 °C (HERAcell, catalog number: HERAcell® 240 )
- Developer (TAEAHN, catalog number: TM-90S )
- Vortex (JEIO TECH, catalog number: VM-96B )
- Deep freezer (Forma Scientific, catalog number: 917 )
- Shaker (SLB, catalog number: SLRM-3 )
Procedure
- mRNA preparation
Phagemid-based K562 cDNA expression libraries were constructed by isolating mRNA from cells with an Ultraspec-RNA isolating system and a biotinylated oligo (dT) probe. The detailed procedure is described below:- Spin down K562 cells (5 ~ 10 x 106) at 5,000 rpm for 5 min at 4 °C.
- Add 1 ml of UltraspecTM RNA reagent to lyse cells by repetitive pipetting. Incubate at 4 °C for 5 min.
- Add 0.2 ml of chloroform and cover the samples tightly, shake vigorously for 15 sec and place on ice at 4 °C for 5 min.
- Centrifuge at 12,000 x g for 15 min at 4 °C.
- Carefully transfer the aqueous phase (about 4/5th volume) to a new fresh tube.
- Add equal volume of isopropanol and store sample for 10 min at 4 °C. Centrifuge at 12,000 x g for 10 min at 4 °C.
- Remove the supernatant and wash RNA pellet twice with 1 ml of 75% ethanol by vortexing and subsequent centrifuge for 5 min at 7,500 x g at 4 °C.
- Remove the supernatant and briefly air dry the RNA pellet for 5 min. Dissolve the RNA pellet in 50-100 ul of Ultraspec DEPC treated water by vortexing for 1 min. Measure RNA concentration using Nanodrop.
- Spin down K562 cells (5 ~ 10 x 106) at 5,000 rpm for 5 min at 4 °C.
- cDNA library construction
Double stranded cDNA was synthesized using a cDNA synthesis kit with an oligo(dT) linker primer, and the resultant cDNA was ligated into the λZAP II express phage vector. The detailed procedure is described below:- Set up a positive control ligation to ligate the test insert into the ZAP Express vector as follows:
- 1 μl of the ZAP Express vector (1 μg)
- 1.6 μl of test insert (0.4 μg)
- 0.5 μl of 10x ligase buffer
- 0.5 μl of 10 mM rATP (pH 7.5)
- 0.9 μl of water
- Then add 0.5 μl of T4 DNA ligase (4 U/μl)
- 1 μl of the ZAP Express vector (1 μg)
- To prepare the sample ligation, add the following components:
- 2 μl of resuspended cDNA (~100 ng)
- 0.5 μl of 10x ligase buffer
- 0.5 μl of 10 mM rATP (pH 7.5)
- 1.0 μl of the ZAP Express vector (1 μg/μl)
- 0.5 μl of water for a final volume of 4.5 μl
- Then add 0.5 μl of T4 DNA ligase (4 U/μl)
- 2 μl of resuspended cDNA (~100 ng)
- Incubate the reaction tubes overnight at 4 °C.
- After ligation is complete, package 1 μl of each ligation using Gigapack III Gold packaging extract in the λZAP II express phage vector package to produce transduced lambda phages according to the packaging instructions outlined in Packaging.
- Set up a positive control ligation to ligate the test insert into the ZAP Express vector as follows:
- Library plating
- Add 1 μl of the lambda phage to 200 μl of XL1-Blue host cells.
- Incubate the phage and bacteria for 15 min at 37 °C to allow the phage to attach to the cells.
- Phagemids (1-1.5 x 104 pfu/plate) were inoculated onto LB plates harboring a lawn of XL1-Blue cells for 4 h incubation at 37 °C.
- Add 1 μl of the lambda phage to 200 μl of XL1-Blue host cells.
- Plaque lifts
- Carefully place the nitrocellulose filter (contain 20 mM IPTG) on top of the plate (not letting any bubbles to form between the nitrocellulose and the plate.
- Leave filter for 10 h on the plate.
- Carefully place the nitrocellulose filter (contain 20 mM IPTG) on top of the plate (not letting any bubbles to form between the nitrocellulose and the plate.
- Prehybridization and Hybridization reactions
- The membranes were blocked in screening buffer containing 0.1 mg/ml yeast RNA in order to reduce nonspecific binding of the probe RNA.
- Specific RNA-protein interactions were detected by hybridization with the [α-32P]-labeled G8-RNA ligand (~ 0.5 x 106 cpm/ml) in screening buffer.
- Let hybridization go 2 h at 4 °C.
- The membranes were blocked in screening buffer containing 0.1 mg/ml yeast RNA in order to reduce nonspecific binding of the probe RNA.
- Washes
- Non-specific bound radioactivity was removed by washing four times at room temperature in 100 ml screening buffer for 5 min each. (Check the membranes for radioactivity using Mini 900 Scintillation monitor.) If it is low (less than 1 count per second), stop here; otherwise, if the signal is still too high (higher than 1 count per second), do the last, stringent, wash).
- Remove membranes and let dry for > 10 min.
- Non-specific bound radioactivity was removed by washing four times at room temperature in 100 ml screening buffer for 5 min each. (Check the membranes for radioactivity using Mini 900 Scintillation monitor.) If it is low (less than 1 count per second), stop here; otherwise, if the signal is still too high (higher than 1 count per second), do the last, stringent, wash).
- Exposures
- Cover membranes with plastic wrap.
- In the dark, place Kodak X-Omat AR film on top of membranes and tape the film to prevent it from moving out of position.
- Leave at -70 °C for 24 h (don't use cassettes without intensifying screens).
- Develop film.
- Cover membranes with plastic wrap.
- Plaque isolation and Clone purification
- Using the developed films, align the plates with the developed film and isolate a positive plaque.
- Place the plug in 1 ml of SM buffer and store at 4 °C.
- Elute the phage from the plug by rocking O/N at 4 °C.
- Repeat screening rounds until a single plaque can be isolated that when spread on a plate and probed will show that all the plaques are positives (usually 3 screens are enough to achieve this).
- Using the developed films, align the plates with the developed film and isolate a positive plaque.
- Excision of plasmids from phage
Each positive phagemid was converted into a plasmid (pBK-CMV) by inoculating the phage into XLOR cells along with the ExAssist helper phage. The detailed procedure is described below:- The plaque of interest from the agar plate and transfer the plaque to a sterile centrifuge tube containing 500 μl of SM buffer and 20 μl of chloroform.
- Vortex the centrifuge tube to release the phage particles into the SM buffer. Incubate the centrifuge tube for overnight at 4 °C.
- Grow separate 50 ml overnight cultures of XL1-Blue and SOLR cells in LB broth with supplements at 30 °C. Gently spin down the XL1-Blue and SOLR cells (1,000 x g). Resuspend each of the cell pellets in 25 ml of 10 mM MgSO4.
- Combine the following components in a 15 ml BD Falcon polypropylene round-bottom tube:
- 200 μl of XL1-Blue cells at an OD600 of 1.0
- 250 ul of phage stock (containing >1 x 105 phage particles)
- 1 μl of the ExAssist helper phage (>1 x 106 pfu/μl)
- 200 μl of XL1-Blue cells at an OD600 of 1.0
- Incubate at 37 °C for 15 min to allow the phage to attach to the cells.
- Add 3 ml of LB broth with supplements and incubate for 2.5-3 h at 37 °C with shaking.
- Heat the tube at 65-70 °C for 20 min to lyse the lambda phage particles and the cells. Spin the tube at 1,000 x g for 15 min to pellet the cell debris.
- Decant the supernatant into a sterile 15 ml tube. This stock contains the excised pBluescript phagemid packaged as filamentous phage particles.
- To plate the extracted phagemids, add 200 μl of freshly grown SOLR cells from step I3 to two 1.5 ml centrifuge tubes. Add 100 μl of the phage supernatant to one centrifuge tube and 10 μl of the phage supernatant to the other centrifuge tube.
- Incubate the centrifuge tubes at 37 °C for 15 min. Plate 200 μl of the cell mixture from each centrifuge tube on LB-ampicillin agar plates (100 μg/ml) and incubate the plates overnight at 37 °C.
- Positive clones were identified by sequencing.
- The plaque of interest from the agar plate and transfer the plaque to a sterile centrifuge tube containing 500 μl of SM buffer and 20 μl of chloroform.
Representative data
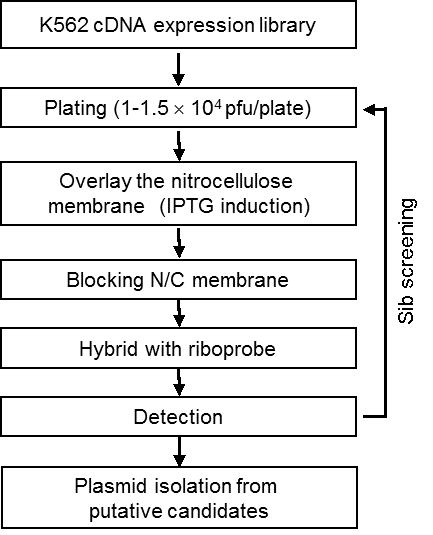
Figure 1. Scheme of RNA-Ligand based cDNA expression library screening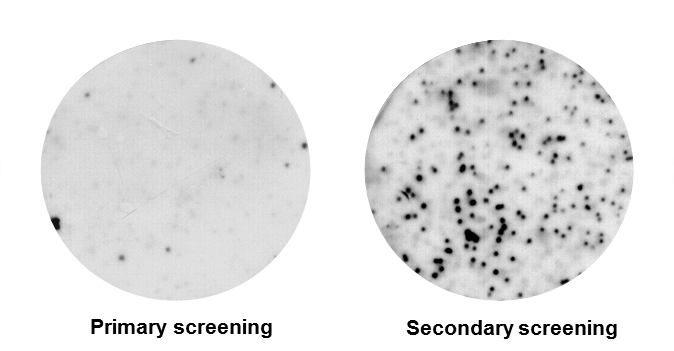
Figure 2. Autoradiograph showing positive putative clones after primary screening. All RNA-binding proteins were additionally confirmed by secondary/tertiary screening.
Notes
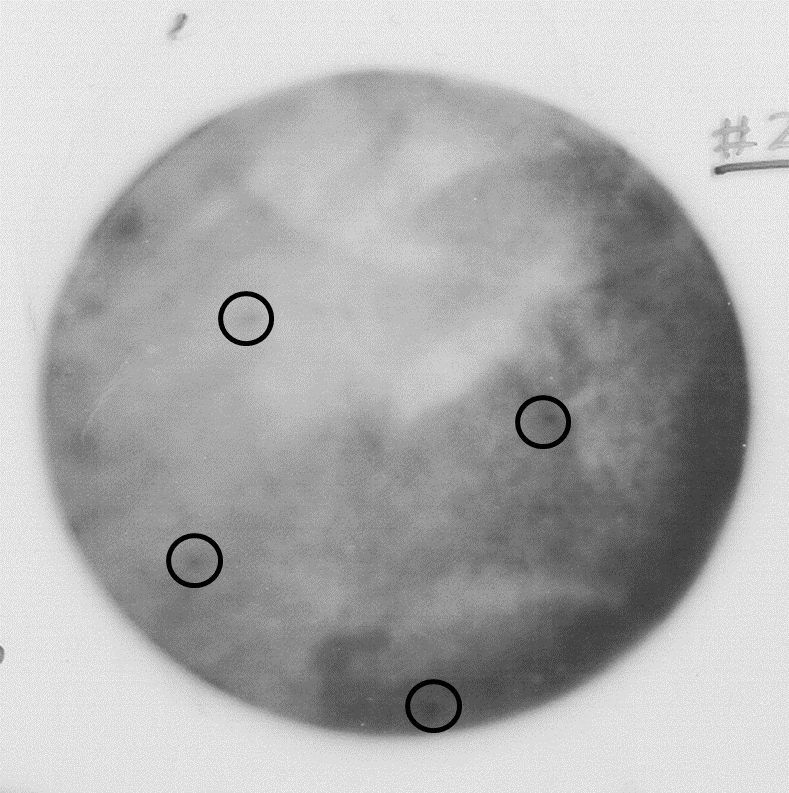
Figure 3. Autoradiography showing high background due to the low concentration of competitor yeast RNA (step E) and poor washing of non-specific bound radioactivity (step F)
Recipes
- Screening buffer
15 mM HEPES (pH 7.9)
50 mM KCl
0.1% (w/v) Ficoll 400-DL
0.01% polyvinyl-pyrolidon PVP-40
0.1 mM MnCl2
0.1 mM ZnCl2
0.1 mM EDTA
0.5 mM DTT - LB plate
1.0 g Tryptone
1.0 g NaCl
0.5 g Yeast Extract
DW 100 ml - SM buffer
5.8 g NaCl
2 g MgSO4
50 mM Tris HCl (pH 7.5)
100 mg Gelatin
DW 1 L
Acknowledgments
This work was supported by Basic Science Research Program (2010-00252250 to C. G. K.), National Research Foundation (NRF), Ministry of Education, Science and Technology (MEST), Republic of Korea. Converging Research Center Program (2013K000283 to C. G. K.), Ministry of Science, ICT & Future Planning (MSIFP), Republic of Korea.
References
- Sagesser, R., Martinez, E., Tsagris, M. and Tabler, M. (1997). Detection and isolation of RNA-binding proteins by RNA-ligand screening of a cDNA expression library. Nucleic Acids Res 25(19): 3816-3822.
- Sanger, F., Nicklen, S. and Coulson, A. R. (1977). DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74(12): 5463-5467.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kim, M. Y., Lee, J. J. and Kim, C. G. (2016). Identification of RNA-binding Proteins by RNA Ligand-based cDNA Expression Library Screening. Bio-protocol 6(2): e1715. DOI: 10.21769/BioProtoc.1715.
Category
Molecular Biology > Protein > Expression
Molecular Biology > RNA > RNA-protein interaction
Biochemistry > Protein > Interaction > Phage display
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












