- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Characterization of HBV Isolates from Patient Serum Samples and Cloning
Published: Vol 5, Iss 24, Dec 20, 2015 DOI: 10.21769/BioProtoc.1683 Views: 10536
Reviewed by: Valeria LullaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Primer Extension Analysis of HBV DNA with Strand-Specific Primers
Jaesung Jung and Kyongmin Kim
Aug 5, 2015 8189 Views

Quantification of Hepatitis B Virus Covalently Closed Circular DNA in Infected Cell Culture Models by Quantitative PCR
Bingqian Qu and Stephan Urban
Apr 5, 2019 8283 Views

Inoculation of Maize with Sugarcane Mosaic Virus Constructs and Application for RNA Interference in Fall Armyworms
Iram Gull and Georg Jander
Jul 20, 2023 2203 Views
Abstract
Hepatitis B virus (HBV) mutants can lead to vaccine failure, diagnostic failure of HBV detection, increase viral replication and resistance to antiviral agents. To study the biological characteristics of these mutations may contribute to our knowledge on viral pathogenesis. Therefore, it is essential to isolate and characterize HBV strains from patients. Here we describe the experimental methods to isolate and clone HBV DNA from patient serum. The method will facilitate isolation and functional analysis of new HBV variants.
Materials and Reagents
- Extraction of HBV DNA from patient serum samples
- 1.5 ml microfuge tube (Axygen, catalog number: 17615044 )
- Patient serum samples
- Tris phenol (Beijing Shuang Xiang Da Company, catalog number: 108952 )
- Trichloromethane (Sinopharm Chemical Reagent Co., catalog number: 61553 )
- Ethanol (Sinopharm Chemical Reagent Co., catalog number: 32061 )
- Isopropyl alcohol (Sinopharm Chemical Reagent Co., catalog number: 32064 )
- Yeast RNA (Life Technologies, Ambion®, catalog number: AM7118 )
Note: Currently, it is “Thermo Fisher Scientific, AmbionTM, catalog number: AM7118”. - Sodium acetate trihydrate (NaOAc) (Sigma-Aldrich, catalog number: 236500 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: EDS )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771 )
- Tris-base (Sigma-Aldrich, catalog number: T1378 )
- Viral lysis buffer (see Recipes)
- 1.5 ml microfuge tube (Axygen, catalog number: 17615044 )
- Amplification of HBV DNA by Polymerase Chain Reaction (PCR)
- KOD-PLUS: a high-fidelity DNA polymerase (TOYOBO CO., catalog number: KOD201 )
- Primers used for PCR as described below (Detailed description can refer to Figure 1)
Name Type Sequence 5’-3’ Position of 5’-base P1 forward CCGGCGTCGACGAGCTCTTCT
TTTTCACCTCTGCCTAATCA1821 P2 forward CCGGCGTCGACGAGCTCTTCA
AAAAGTTGCATGGTGCTGG1825 P3 reverse CACTGAACAAATGGCACTAGT
AAACTGAGCC699 P4 reverse GGCTCAGTTTACTAGTGCCATT
TGTTCAGTG669
Note: The underlined red letters are the restriction endonuclease binding site. P1: Sal I, Sac I; P2: Sal I, Sac I; P3: Spe I; P4: Spe I. P1/P3 are used to amplify the 2.05 kb fragment (1,821-699 bp), P2/P4 are used to amplify the 1.15 kb fragment (669-1,825 bp).
- KOD-PLUS: a high-fidelity DNA polymerase (TOYOBO CO., catalog number: KOD201 )
- Gel extraction of HBV DNA
- AxyPrepTM DNA Gel Extraction kit (Thermo Fisher Scientific, AxygenTM, catalog number: APGX50 )
- Add dATP to generate 3’ overhangs
- Taq DNA polymerase (TOYOBO CO., catalog number: TAP201 )
- dATP (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R0141 )
- Taq DNA polymerase (TOYOBO CO., catalog number: TAP201 )
- Integration of HBV DNA into pGEM®-T Vector
- pGEM®-T Vector System I (Promega Corporation, catalog number: A3600 )
- 0.5 ml microfuge tube (Axygen, catalog number: 05615119 )
- pGEM®-T Vector System I (Promega Corporation, catalog number: A3600 )
- Transformation
- Competent DH5α (Takara, catalog number: 9057 )
- Ampicillin (Sinopharm Chemical Reagent Co., catalog number: 69523 )
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, catalog number: PHG0010-5G )
- 5-Bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal) (Sigma-Aldrich, catalog number: B4252-50MG )
- Sodium chloride (NaCl) (Sinopharm Chemical Reagent CO., catalog number: 10019318 )
- TRYPTONE (Oxoid Limited, catalog number: LP0042 )
- YEAST EXTRACT (Oxoid Limited, catalog number: LP0021 )
- LB medium (see Recipes)
- Competent DH5α (Takara, catalog number: 9057 )
- Extract HBV DNA-integrated plasmid from E.coli-DH5α
- E.Z.N.A.® Plasmid DNA Mini Kit I (Omega Bio-tek Inc., catalog number: D6942-02 )
- Integration of full-length HBV genome into the cloning vector pUC19
Equipment
- Instrument for PCR (Biocompare, Biometra, catalog number: 070851 )
- Centrifuge (Eppendorf, model: 5424 )
- Electrophoresis system (Beijing Liuyi Biotechnology, model: DYY6C )
- Electronic scales (Mettler-Toledo International Inc., model: PB602-N )
Note: Currently, it is “Mettler-Toledo International Inc., model: PB602-S ”. - Bacteriological incubator (bio-equip, Shanghai Jing Hong Laboratory Instrument Co., model number: GNP-9080 )
- BIO IMAGING SYSTEM (Syngene, model: SYDR2/1361 )
- Heat block
Procedure
- Extraction of HBV DNA from patient serum samples
- Add 100 μl viral lysis buffer to 100 μl serum samples, incubate at 65 °C for 4 h.
- Add 125 μl Tris phenol and 125 μl Trichloromethane, vigorously vortex for 10 sec. Then, centrifuge at 13,000 rpm for 5 min at room temperature. This step can separate protein from nucleic acid and nucleic acid would be in the aqueous upper layer.
- Transfer aqueous upper layer into a new 1.5 ml tube.
- Add 20 μg Yeast RNA (as a co-precipitant here), 0.1 volume NaOAc and 0.7 volume Isopropyl alcohol. After inverting several times, precipitate the mixture overnight at -20 °C.
- Centrifuge at 13,000 rpm for 15 min at 4 °C and throw away the supernatant.
- Add 1 ml of 70% ethanol and gently invert.
- Centrifuge at 13,000 rpm for 5 min at 4 °C and throw away the supernatant.
- Dry the DNA pellet for 5~10 min (air-drying) and dissolve in 20 μl ddH2O as the PCR template.
- Add 100 μl viral lysis buffer to 100 μl serum samples, incubate at 65 °C for 4 h.
- Amplification of HBV DNA by Polymerase Chain Reaction (PCR)
- PCR system:
Reagent Volume/μl 10x KOD buffer 5 μl dNTPs (2.5 mM each) 5 μl P1 (20 mM) 1 μl P3 (20 mM) 1 μl Template 5 μl Mg2+ (25 mM) 2 μl KOD_PLUS enzyme (1 U/μl) 1 μl
Add ddH2O to a final volume of 50 μl. - PCR reaction program:
94 °C, 5 min
94 °C, 30 sec
55 °C, 30 sec 40 cycles
68 °C, 2 min
68 °C, 10 min
For the 1.15 kb fragment amplification, P2 and P4 primers are used and the extension proceeding of PCR reaction program should be changed from 68 °C, 2 min to 68 °C, 1 min. - Add 10x DNA loading buffer to each sample and mix by pipetting. Purifying the PCR product by running on a 1% agarose gel at 100 V for 30 min.
Note: The size of gel used here is 6 cm; the voltage/run-time would vary by the gel rig used.
- PCR system:
- Gel extraction of HBV DNA
- Excise the agarose gel slice containing the DNA fragment of interest with a clean, sharp scalpel under ultraviolet illumination. Briefly place the excised gel slice on absorbent toweling to remove residual buffer. Transfer the gel slice to a piece or plastic wrap or a weighing boat. Mince the gel into small pieces and weigh. In this application, the weight of gel is regarded as equivalent to the volume. For example, 100 mg of gel is equivalent to a 100 μl volume. Transfer the gel slice into a 1.5 ml microfuge tube.
Note: Alternatively, the gel slice can be placed into the 1.5 ml microfuge tube and then crushed with a pipette tip or other suitable device. Spin the tube for 30 sec at 12,000 x g to consolidate the gel at the bottom of the tube. Use the graduations to estimate the volume of the agarose gel. - Add a 3x sample volume of Buffer DE-A.
Note: The color of Buffer DE-A is red. This color is used to add contrast in the next step, so that any pieces of unsolubilized agarose can be visualized. - Resuspend the gel in Buffer DE-A by vortexing. Heat at 75 °C until the gel is completely dissolved (typically, 6-8 min). Heat at 40 °C if low-melt agarose gel is used. Intermittent vortexing (every 2-3 min) will accelerate gel solubilization.
IMPORTANT: Gel must be completely dissolved or the DNA fragment recovery will be reduced.
IMPORTANT: Do not heat the gel for longer than 10 min. - Add 0.5x Buffer DE-A volume of Buffer DE-B, mix. If the DNA fragment is less than 400 bp, supplement further with a 1x sample volume of isopropanol.
Example: For a 1% gel slice equivalent to 100 μl, add the following:
• 300 μl Buffer DE-A
• 150 μl Buffer DE-B
If the DNA fragment is <400 bp, you would also add:
• 100 μl of isopropanol
Note: The color of the mixture will turn yellow after the addition of Buffer DE-B. Please make sure the contents are a uniform yellow color before proceeding. - Place a Miniprep column into a 2 ml microfuge tube (provided). Transfer the solubilized agarose from step C4 into the column. Centrifuge at 12,000 x g for 1 min.
- Discard the filtrate from the 2 ml microfuge tube. Return the Miniprep column to the 2 ml microfuge tube and add 500 μl of Buffer W1. Centrifuge at 12,000 x g for 30 sec.
- Discard the filtrate from the 2 ml microfuge tube. Return the Miniprep column to the 2 ml microfuge tube and add 700 μl of Buffer W2. Centrifuge at 12,000 x g for 30 sec.
Note: Make sure that 95-100% ethanol has been added into Buffer W2 concentrate. Make a notation on the bottle label for future reference. - Discard the filtrate from the 2 ml microfuge tube. Place the Miniprep column back into the 2 ml microfuge tube. Add a second 700 μl aliquot of Buffer W2 and centrifuge at 12,000 x g for 1 min.
Note: Two washes with Buffer W2 are used to ensure the complete removal of salt, eliminating potential problems in subsequent enzymatic reactions, such as ligation and sequencing reaction. - Discard the filtrate from the 2 ml microfuge tube. Place the Miniprep column back into the 2 ml microfuge tube. Centrifuge at 12,000 x g for 1 min.
- Transfer the Miniprep column into a clean 1.5 ml microfuge tube (provided). To elute the DNA, add 25-30 μl of Eluent or deionized water to the center of the membrane. Let it stand for 1 min at room temperature. Centrifuge at 12,000 x g for 1 min.
Notes:- Pre-warming the Eluent at 65 °C will generally improve elution efficiency.
- Deionized water can also be used to elute the DNA fragments.
- More detailed information can refer to the PDF protocol file attached here.
- Pre-warming the Eluent at 65 °C will generally improve elution efficiency.
- Excise the agarose gel slice containing the DNA fragment of interest with a clean, sharp scalpel under ultraviolet illumination. Briefly place the excised gel slice on absorbent toweling to remove residual buffer. Transfer the gel slice to a piece or plastic wrap or a weighing boat. Mince the gel into small pieces and weigh. In this application, the weight of gel is regarded as equivalent to the volume. For example, 100 mg of gel is equivalent to a 100 μl volume. Transfer the gel slice into a 1.5 ml microfuge tube.
- Add dATP to generate 3’ overhangs
- The reaction system to add dATP:
Reagent Volume/μl dATP 1 μl 10x Taq buffer 1 μl Taq 1 μl PCR product 7 μl - Incubate at 72 °C for 30 min.
- The reaction system to add dATP:
- Integration of HBV DNA into pGEM®-T Vector
- Briefly centrifuge the pGEM®-T Vector tube to collect contents at the bottom of the tube.
- Set up ligation reactions as described below. Vortex the 2x Rapid Ligation Buffer vigorously before each use. Use 0.5 ml tubes known to have low DNA-binding capacity.
Reagents Volume 2X Rapid Ligation Buffer 5 μl pGEM®-T Vector (50 ng) 1 μl PCR product 2 μl T4 DNA Ligase (3 Weiss units/µl) 1 μl Deionized water to a final volume of 10 μl - Mix the reactions by pipetting. Incubate the reactions 1 h at room temperature. Alternatively, incubate the reactions overnight at 4 °C for the maximum number of transformants.
Note: More detailed information can refer to the PDF protocol file attached here.
- Briefly centrifuge the pGEM®-T Vector tube to collect contents at the bottom of the tube.
- Transformation
- Add 5 μl ligation product into 50 μl competent E.coli-DH5α, incubate on ice for 30 min.
- Incubate at 42 °C for 90 sec followed by transfering immediately on ice for 3 min.
- Add 1 ml LB medium, incubate at 37 °C shaker for 1 h.
- Centrifuge at 8,000 rpm for 2 min at room temperature and throw away the supernatant, use 50 μl LB medium to re-suspend the bacteria precipitate.
- Add 2 μl IPTG (storage concentration: 20%, m/V) and 20 μl X-gal (storage concentration: 2%, m/V) and mix by pipetting, then spread the solution on Amp resistance plate (final concentration of Amp: 50 μg/ml).
- Put the plate at 37 °C bacteriological incubator for 12-16 h.
- Choose the white clones to get the HBV DNA-integrated plasmid.
- Add 5 μl ligation product into 50 μl competent E.coli-DH5α, incubate on ice for 30 min.
- Extract HBV DNA-integrated plasmid from E.coli-DH5α
- Isolate a single white colony from the plate (step F6), and inoculate a culture of 5 ml LB medium containing the Amp antibiotic. Incubate for ~12-16 h at 37 °C with vigorous shaking (~ 300 rpm). Use a 10-20 ml culture tube or a flask with a volume of at least 4 times the volume of the culture.
- Centrifuge at 10,000 x g for 1 min at room temperature.
- Decant or aspirate and discard the culture media.
- Add 250 μl Solution I/RNase A. Vortex or pipet up and down to mix thoroughly. Complete resuspension of cell pellet is vital for obtaining good yields.
Note: RNase A must be added to Solution I before use. Please see the instructions (attached here) in the Preparing Reagents section on Page 6. - Transfer suspension into a new 1.5 ml microcentrifuge tube.
- Add 250 μl Solution II. Invert and gently rotate the tube several times to obtain a clear lysate. A 2-3 min incubation may be necessary.
Note: Avoid vigorous mixing as this will shear chromosomal DNA and lower plasmid purity. Do not allow the lysis reaction to proceed more than 5 min. Store Solution II tightly capped when not in use to avoid acidification from CO2 in the air. - Add 350 μl Solution III. Immediately invert several times until a flocculent white precipitate forms.
Note: It is vital that the solution is mixed thoroughly and immediately after the addition of Solution III to avoid localized precipitation. - Centrifuge at maximum speed (≥13,000 x g) for 10 min. A compact white pellet will form. Promptly proceed to the next step.
- Insert a HiBind® DNA Mini Column into a 2 ml Collection Tube.
- Transfer the cleared supernatant from step G8 by CAREFULLY aspirating it into the HiBind® DNA Mini Column. Be careful not to disturb the pellet and that no cellular debris is transferred to the HiBind® DNA Mini Column.
- Centrifuge at maximum speed for 1 min.
- Discard the filtrate and reuse the collection tube.
- Add 500 μl HB Buffer.
- Centrifuge at maximum speed for 1 min.
- Discard the filtrate and reuse collection tube.
- Add 700 μl DNA Wash Buffer.
Note: DNA Wash Buffer must be diluted with 100% ethanol prior to use. Please see Page 6 (attached here) for instructions. - Centrifuge at maximum speed for 1 min.
- Discard the filtrate and reuse the collection tube.
- Centrifuge the empty HiBind® DNA Mini Column for 2 min at maximum speed to dry the column matrix.
Note: It is important to dry the HiBind® DNA Mini Column matrix before elution. Residual ethanol may interfere with downstream applications. - Transfer the HiBind® DNA Mini Column to a clean 1.5 ml microcentrifuge tube.
- Add 30-100 μl Elution Buffer or sterile deionized water directly to the center of the column membrane.
Note: The efficiency of eluting DNA from the HiBind® DNA Mini Column is dependent on pH. If using sterile deionized water, make sure that the pH is around 8.5. - Let sit at room temperature for 1 min.
- Centrifuge at maximum speed for 1 min.
Note: This represents approximately 70% of bound DNA. An optional second elution will yield any residual DNA, though at a lower concentration. - Plasmid sequencing and choose the right HBV DNA-integrated plasmid for below step.
Note: More detailed information can refer to the PDF protocol file attached here.
- Isolate a single white colony from the plate (step F6), and inoculate a culture of 5 ml LB medium containing the Amp antibiotic. Incubate for ~12-16 h at 37 °C with vigorous shaking (~ 300 rpm). Use a 10-20 ml culture tube or a flask with a volume of at least 4 times the volume of the culture.
- Integration of full-length HBV genome into the cloning vector pUC19
- Digest the 2.05 kb-HBV DNA-integrated plasmid with Sal I and Spe I, 1.15-HBV DNA-integrated plasmid with Sac I and Spe I, pUC19 plasmid with Sal I and Sac I. The detailed reaction system as described below.
Reagents 2.05 kb-HBV DNA- integrated plasmid 1.15 kb-HBV DNA- integrated plasmid pUC19 10x Digest buffer 2 μl 2 μl 2 μl Sal I 1 μl - 1 μl Spe I 1 μl 1 μl - Sac I - 1 μl 1 μl Plasmid 1 μg 1 μg 1 μg Deionized water to a final volume of 20 μl 20 μl 20 μl - Incubate at 37 °C for 1 h.
- Add 10x DNA loading buffer to each sample and mix by pipetting. Gel purify the product by running on a 1% agarose gel at 100 V for 30 min.
- Gel extraction of the 2.05 kb fragment, 1.15 kb fragment and ~2.7 kb linear pUC19 fragment as described in step C.
- Use T4 DNA ligase to integrate them at 4 °C overnight. The detailed reaction system as described below.
Reagents Volume 10x Ligation Buffer 1 μl ~2.7 kb linear pUC19 fragment 1 μl 2.05 kb fragment 3 μl 1.15 kb fragment 4 μl T4 DNA Ligase 1 μl a final volume of 10 μl - Incubate the reactions overnight at 4 °C for the maximum number of transformants.
- Ligation products are transformed into the competent E.coli-DH5α as described in step F1-6.
- Extract recombinant plasmid from E.coli-DH5α as described step G.
- Identify the plasmid with Sac I and Spe I. The detailed steps can refer to H1-3.
- The plasmid that three fragments: 2.7 kb, 2.05 kb and 1.15 kb formed by digestion with Sac I and Spe I (Figure 2C) is the right full-length HBV genome-integrated plasmid, pUC/HBV plasmid.
- Digest the 2.05 kb-HBV DNA-integrated plasmid with Sal I and Spe I, 1.15-HBV DNA-integrated plasmid with Sac I and Spe I, pUC19 plasmid with Sal I and Sac I. The detailed reaction system as described below.
Representative data
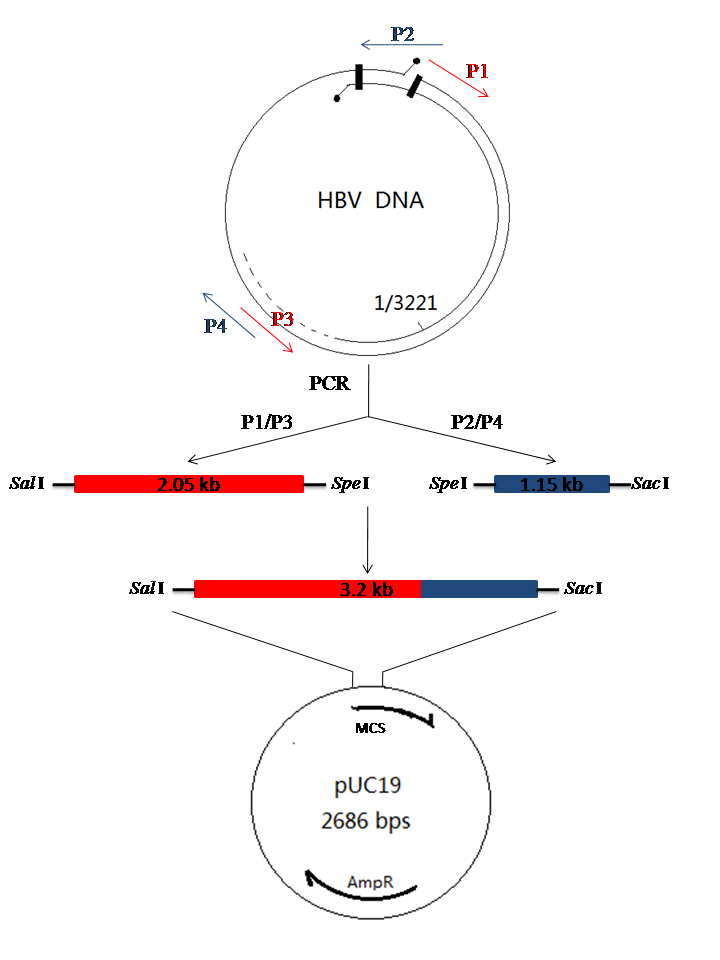
Figure 1. Schematic diagram of cloning for HBV DNA. HBV DNA was extracted from patient serum as the PCR template. Primer P1 and P3 are used to amplify the 2.05 kb-HBV DNA fragment; Primer P2 and P4 are used to amplify the 1.15 kb-HBV DNA fragment. The two fragments were ligated into pGEM®-T Vector, respectively and subjected to sequence analysis. The 2.05 kb-HBV DNA fragment and 1.15 kb-HBV DNA fragment were digested from the right recombinant plasmid and were ligated into the pUC19 plasmid to form the recombinant pUC/HBV plasmid.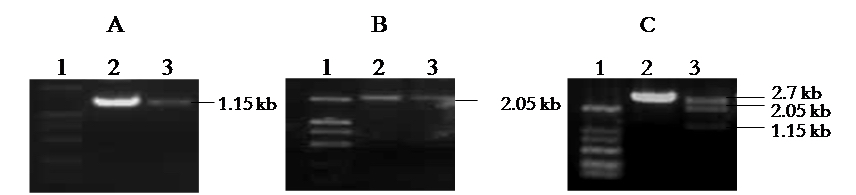
Figure 2. Construction and Identification of pUC/HBV. A. Amplification of 1.15 kb HBV DNA fragment. Lane 1: Marker; Lane 2,3: samples. B. Amplification of 2.05 kb HBV DNA fragment. Lane 1: Marker; Lane 2,3: samples. C. Identification of pUC/HBV by Sac I and Spe I digestion. Lane 1: Marker; Lane 2: negative clone; Lane 3: positive clone.
Recipes
- Viral lysis buffer
20 mM Tris-HCl (pH 8.0)
10 mM EDTA
0.1% SDS
0.8 mg/ml proteinase K - LB medium
10 g/L NaCl
10 g/L tryptone
5 g/L yeast extract
Acknowledgments
This protocol was modified from the previous work by Hui Shi and Liang Cao. This study was supported by the National Nature Science Foundation of China (31200699).
References
- Alcantara, F. F., Tang, H. and McLachlan, A. (2002). Functional characterization of the interferon regulatory element in the enhancer 1 region of the hepatitis B virus genome. Nucleic Acids Res 30(9): 2068-2075.
- Cao, L., Wu, C., Shi, H., Gong, Z., Zhang, E., Wang, H., Zhao, K., Liu, S., Li, S., Gao, X., Wang, Y., Pei, R., Lu, M. and Chen, X. (2014). Coexistence of hepatitis B virus quasispecies enhances viral replication and the ability to induce host antibody and cellular immune responses. J Virol 88(15): 8656-8666.
- Gunther, S., Li, B. C., Miska, S., Kruger, D. H., Meisel, H. and Will, H. (1995). A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol 69(9): 5437-5444.
- He, J., He, L. and Yao, X. (1998). A novel method to study the full length genome of hepatitis B virus. Chin J Infect Dis 16(2): p70-72.
- Huang, L. R., Wu, H. L., Chen, P. J. and Chen, D. S. (2006). An immunocompetent mouse model for the tolerance of human chronic hepatitis B virus infection. Proc Natl Acad Sci U S A 103(47): 17862-17867.
- Li, L., Shen, H., Li, A., Zhang, Z., Wang, B., Wang, J., Zheng, X., Wu, J., Yang, D., Lu, M. and Song, J. (2013). Inhibition of hepatitis B virus (HBV) gene expression and replication by HBx gene silencing in a hydrodynamic injection mouse model with a new clone of HBV genotype B. Virol J 10: 214.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cao, L., Wu, C., Zhao, K. and Chen, X. (2015). Characterization of HBV Isolates from Patient Serum Samples and Cloning. Bio-protocol 5(24): e1683. DOI: 10.21769/BioProtoc.1683.
Category
Microbiology > Microbial biochemistry > DNA
Molecular Biology > DNA > DNA cloning
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









