- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Telomerase Repeated Amplification Protocol (TRAP)
Published: Vol 5, Iss 22, Nov 20, 2015 DOI: 10.21769/BioProtoc.1657 Views: 23168
Reviewed by: HongLok LungVanesa Olivares-IllanaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Telomere Dysfunction Induced Foci (TIF) Analysis
Ilgen Mender and Jerry W. Shay
Nov 20, 2015 13128 Views

Telomere Restriction Fragment (TRF) Analysis
Ilgen Mender and Jerry W. Shay
Nov 20, 2015 22134 Views
Abstract
Telomeres are found at the end of eukaryotic linear chromosomes, and proteins that bind to telomeres protect DNA from being recognized as double-strand breaks thus preventing end-to-end fusions (Griffith et al., 1999). However, due to the end replication problem and other factors such as oxidative damage, the limited life span of cultured cells (Hayflick limit) results in progressive shortening of these protective structures (Hayflick and Moorhead, 1961; Olovnikov, 1973). The ribonucleoprotein enzyme complex telomerase- consisting of a protein catalytic component hTERT and a functional RNA component hTR or hTERC- counteracts telomere shortening by adding telomeric repeats to the end of chromosomes in ~90% of primary human tumors and in some transiently proliferating stem-like cells (Shay and Wright, 1996; Shay and Wright, 2001). This results in continuous proliferation of cells which is a hallmark of cancer. Therefore, telomere biology has a central role in aging, cancer progression/metastasis as well as targeted cancer therapies. There are commonly used methods in telomere biology such as Telomere Restriction Fragment (TRF) (Mender and Shay, 2015b), Telomere Repeat Amplification Protocol (TRAP) and Telomere dysfunction Induced Foci (TIF) analysis (Mender and Shay, 2015a). In this detailed protocol we describe Telomere Repeat Amplification Protocol (TRAP).
The TRAP assay is a popular method to determine telomerase activity in mammalian cells and tissue samples (Kim et al., 1994). The TRAP assay includes three steps: extension, amplification, and detection of telomerase products. In the extension step, telomeric repeats are added to the telomerase substrate (which is actually a non-telomeric oligonucleotide, TS) by telomerase. In the amplification step, the extension products are amplified by the polymerase chain reaction (PCR) using specific primers (TS upstream primer and ACX downstream primer) and in the detection step, the presence or absence of telomerase is analyzed by electrophoresis. TSNT is, an internal standard control, amplified by TS primer. NT is its own reverse primer, which is not a substrate for telomerase. These primers are used to identify false-negative results by if the gel lacks internal control bands.
Materials and Reagents
- Cancer cells (H1299 non-small cell lung, A549 non-small cell lung cancer cells)
- Tris-HCl (pH 8.3 and pH 8.0)
- Magnesium Chloride (MgCl2) (Thermo Fisher Scientific, catalog number: BP241 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9541 )
- TweenTM 20 (Thermo Fisher Scientific, catalog number: BP337 )
- Ethylene glycol-bis(2-aminoethylether)-N, N, N′, N′-tetraacetic acid EGTA (Sigma-Aldrich, catalog number: E3889 )
- Ethylenediaminetetraacetic Acid (EDTA) (Thermo Fisher Scientific, catalog number: BP-120 )
- Nonidet-P40 (Fluka BioChemika, catalog number: 74385 )
Note: Currently, it is “Sigma-Aldrich, catalog number: 74385 ”. - Glycerol (Sigma-Aldrich, catalog number: G5516 )
- 2-mercaptoethanol (Sigma-Aldrich, catalog number: M3148 )
- 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) (Sigma-Aldrich, catalog number: A8456 )
- dNTP (Invitrogen)
- Cy5-TS primer (Sigma-Aldrich)
- ACX (reverse primer) (Sigma-Aldrich)
- TSNT (36-bp internal standard control) (Integrated DNA Technologies)
- NT (reverse primer for internal standard) (Sigma-Aldrich)
- Takara Taq DNA polymerase, hot start version (Takara Bio Company, ClonTech, catalog number: R007A )
- 40% acrylamide and bis-acrylamide solution (19:1) (Bio-Rad Laboratories, catalog number: 161-0144 )
- Ammonium persulfate (APS) (Bio-Rad Laboratories, catalog number: 161-0700 )
- N, N, N’, N’-Tetramethylethylenediamine (TEMED) (Sigma-Aldrich, catalog number: T9281 )
- Primer mix (see Recipes)
- Cy5-TS primer (see Recipes)
- 10x TRAP reaction buffer (see Recipes)
- 50x dNTP (see Recipes)
- NP-40 lysis buffer (see Recipes)
Equipment
- Polymerase Chain Reaction (PCR) Thermo Cycler (Bio-Rad Laboratories, model: PTC-1148 )
- Typhoon PhosphorImager® scanner system (Amersham Biosciences, GE Healthcare, model: Typhoon TRIO )
Software
- ImageQuant Software (Molecular Dynamics)
Procedure
- Prepare cell lysates
- Collect 100,000 cells into a DNase/RNase -free microfuge tube.
- Centrifuge cells at 3,000 x g for 5 min.
- Remove the supernatant.
Note: The pellet doesn’t need to be washed. If samples are to be used at a later time point, put in -80 °C. - Resuspend cell pellet on ice-cold NP-40 lysis buffer at a concentration of 2,500 cells per μl (40 μl lysis buffer for 100,000 cells).
- Leave on ice for 30 min or snap-freeze cell lysates in liquid nitrogen and then place at -80 °C.
- Collect 100,000 cells into a DNase/RNase -free microfuge tube.
- Preparation for PCR-Cy5 fluorescent gel-based TRAP
- Keep samples on ice.
- Choose a positive control (i.e. HCT116, H1299) and prepare three different serial dilutions (1:10) in NP-40.
- Prepare the TRAP master mix. Final volume of the PCR reaction is 50 μl.
Master mix for one sample:
H2O 40.2 μl Trap buffer 5 μl dNTP 1 μl Cy5-TS primer 1 μl Primer mix 1 μl BSA 0.4 μl Taq polymerase 0.4 μl - Add 1 μl of sample to 49 μl of the master mix.
Note: For negative control, 1 μl NP-40 to 49 μl of the master mix.
- Keep samples on ice.
- Amplification of the extension products by PCR
- 25 °C for 40 min for extension.
- 95 °C for 5 min to deactivate telomerase.
- 24 to 29 cycles at:
95 °C for 30 sec
52 °C for 30 sec
72 °C for 45 sec - 72 °C for 10 min
- 4 °C for up to five days
Note: Samples can be stored at -20 °C for 1 month.
- 25 °C for 40 min for extension.
- Running acrylamide gel
- Add 5 μl of loading dye to each sample.
- Making the 10% nondenaturing acrylamide gel.
32.5 ml H2O (MilliQ® water)
12.5 ml 40% acrylamide (19:1 acrylamide:bisacrylamide)
5 ml 5x TBE
250 μl 10% APS
50 μl TEMED - 0.5x TBE running buffer
- Running time is about 2.5 h at 200/220 volts.
- Load 25 μl of the sample to each well.
- Visualize using Typhoon® that can read Cy5 fluorescein.
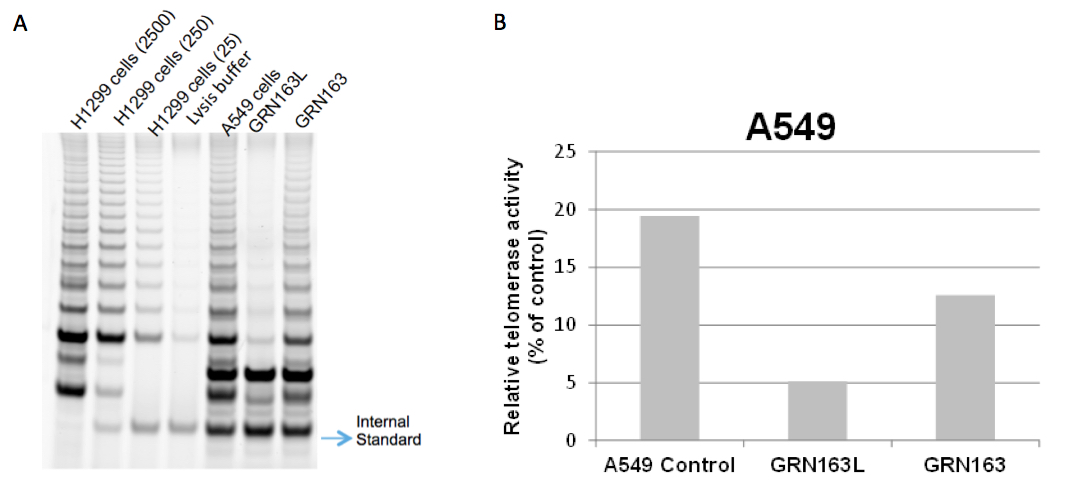
Figure 1. TRAP gels show partial telomerase inhibition with 10 μM GRN163L in A549 non small cell lung cancer cells for 72 h. It is known that a lipid modified N3→P5 thio-phosphoramidate oligonucleotide, GRN163L (also referred to as Imetelstat), inhibits telomerase more potently than its parental non-conjugated thio-phospharamidate sequence, GRN163, in cancer cells (Herbert et al., 2005). This figure also shows that GRN163L (L= lipidated with palmitoyl C16) inhibits A549 cells more potently compared to GRN163. H1299 non-small lung cancer cells were used with three different dilutions as a positive control. 2500, 250 and 25 cells for H1299 were used in lane 1, lane 2 and lane 3, respectively. No cells were added in lysis buffer in lane 4, which is a negative control (A). ImageQuant Software was used to determine the intensity of the telomerase products (6 bp-ladder) and the ITAS (Internal Standard Control, 36 bp) band. The ratio of intensity levels of TRAP sample ladders and ITAS band (intensity of sample’s TRAP ladder/intensity of ITAS band) was calculated and each sample was normalized to the positive control as a percentage (positive control; H1299 2500 cells) after background subtraction (B).
- Add 5 μl of loading dye to each sample.
Recipes
- Primer mix
Primer mix (50X) includes the reverse primer (ACX), the substrate for 36-bp internal standard control (TSNT), and the reverse primer for the internal standard (NT) or ITAS (internal telomerase activity standard).
Sequences: Notes:
Notes:Stock Final concentration ACX 1.0 μg/μl 100 ng/μl NT 1.0 μg/μl 100 ng/μl TSNT 1.0 attomol/μl 0.01 attomol/μl - It is highly recommended to purchase TSNT oligo from a company other than where the ACX and NT primers are bought. This helps prevention of the contamination of TSNT with ACX and NT.
- Preparation of stock TSNT should be done in a separate room from the TRAP bench and use a pipetman that is not used in the TRAP area.
- An attomol is 10-18 mol.
Preparation of stock TSNT:
Dilute the dry TSNT oligonucleotide to 100 μM concentration with DEPC treated water. Then prepare the serial dilutions (1:1,000, 1:1,000, 1:100); 100 nM, 100 pM and 1 pM. So, the final concentration of the TSNT stock will contain a 100x stock of TSNT (1.0 attomol/μl).
Note: To get the correct ITAS signal, you can adjust the amount of 100x stock of TSNT. For example, try a variety of dilutions of 100x stock of TSNT such as 1,000x, 300x, 100x, and 30x and see how the ITAS looks on TRAP ladder.
Preparation of primer mix:- Mix ACX and NT primers together with water.
- Move to the area where the TSNT was prepared and add the TSNT to the mix.
- Clean the outside of the tubes and rack with diluted bleach (spray 10% bleach on the tubes, then dry them with paper towel or spray 10% bleach on paper towel and clean the outside of the tubes).
- Return to the TRAP area with the tube and prepare aliquots to store at -20 °C up to 1 year.
- It is highly recommended to purchase TSNT oligo from a company other than where the ACX and NT primers are bought. This helps prevention of the contamination of TSNT with ACX and NT.
- Cy5-TS primer
TS oligo is purchased modified with Cy5 on the 5’end (HPLC or PAGE purified). Final concentration of TS primer is 100 ng/μl (diluted in DEPC water).
Sequence: 5’-AAT CCG TCG AGC AGA GTT-3’ - 10x TRAP reaction buffer
Final concentration Tris-HCl (pH 8.3) 200 mM MgCl2 15 mM KCl 630 mM Tween 20 0.5% (v/v) EGTA 10 mM - 50x dNTP
Use a mix containing all four DNA nucleotides (dTTP, dATP, dCTP, dGTP) at an equivalent and final concentration of 2.5 mM - NP-40 lysis buffer
Final concentration Tris-HCl (pH 8.0) 10 mM MgCl2 1 mM EDTA 1 mM NP-40 1% (v/v) Sodium deoxycholate 0.25 mM Glycerol 10% (v/v) NaCl 150 mM 2-mercaptoethanol 5 mM AEBSF 0.1 mM
Acknowledgments
Some of these protocols were adapted from previously published studies. Some of TRAP protocols are referenced here (Herbert et al., 2003; Norton et al., 1998; Wright et al., 1995). We thank Zeliha Gunnur Dikmen for her help in acquisition of TRAP gel and Abhijit Bugde from the Live Cell Imaging Facility at UT Southwestern for his assistance with the imaging and analysis part of Telomere dysfunction Induced Foci (TIF) analysis.
References
- Griffith, J. D., Comeau, L., Rosenfield, S., Stansel, R. M., Bianchi, A., Moss, H. and de Lange, T. (1999). Mammalian telomeres end in a large duplex loop. Cell 97(4): 503-514.
- Hayflick, L. and Moorhead, P. S. (1961). The serial cultivation of human diploid cell strains. Exp Cell Res 25: 585-621.
- Olovnikov, A. M. (1973). A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol 41(1): 181-190.
- Shay, J. W. and Wright, W. E. (1996). Telomerase activity in human cancer. Curr Opin Oncol 8(1): 66-71.
- Shay, J. W. and Wright, W. E. (2001). Telomeres and telomerase: implications for cancer and aging. Radiat Res 155(1 Pt 2): 188-193.
- Kim, N. W., Piatyszek, M. A., Prowse, K. R., Harley, C. B., West, M. D., Ho, P. L., Coviello, G. M., Wright, W. E., Weinrich, S. L. and Shay, J. W. (1994). Specific association of human telomerase activity with immortal cells and cancer. Science 266(5193): 2011-2015.
- Herbert, B. S., Gellert, G. C., Hochreiter, A., Pongracz, K., Wright, W. E., Zielinska, D., Chin, A. C., Harley, C. B., Shay, J. W. and Gryaznov, S. M. (2005). Lipid modification of GRN163, an N3'-->P5' thio-phosphoramidate oligonucleotide, enhances the potency of telomerase inhibition. Oncogene 24(33): 5262-5268.
- Herbert, B. S., Shay, J. W. and Wright, W. E. (2003). Analysis of telomeres and telomerase. Curr Protoc Cell Biol Chapter 18: Unit 18 16.
- Mender, I. and Shay, J. W. (2015a). Telomere dysfunction induced foci (TIF) analysis. Bio-protocol 5(22): e1656.
- Mender, I. and Shay, J. W. (2015b). Telomere restriction fragment (TRF) analysis. Bio-protocol 5(22): e1658.
- Norton, J. C., Holt, S. E., Wright, W. E. and Shay, J. W. (1998). Enhanced detection of human telomerase activity. DNA Cell Biol 17(3): 217-219.
- Wright, W. E., Shay, J. W. and Piatyszek, M. A. (1995). Modifications of a telomeric repeat amplification protocol (TRAP) result in increased reliability, linearity and sensitivity. Nucleic Acids Res 23(18): 3794-3795.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Mender, I. and Shay, J. W. (2015). Telomerase Repeated Amplification Protocol (TRAP). Bio-protocol 5(22): e1657. DOI: 10.21769/BioProtoc.1657.
Category
Cancer Biology > Replicative immortality > Cell biology assays > Nucleic acid sequencing
Cancer Biology > Replicative immortality > Cell biology assays > Protein
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












