- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
[14C]-Tryptophan Metabolic Tracing in Liver Cancer Cells
Published: Vol 5, Iss 17, Sep 5, 2015 DOI: 10.21769/BioProtoc.1582 Views: 9914
Reviewed by: Jia LiShannon RuppertYong Teng

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Enhanced Ribonucleoprotein Immunoprecipitation (RIP) Technique for the Identification of mRNA Species in Ribonucleoprotein Complexes
Saja A. Fakhraldeen [...] Caroline M. Alexander
Oct 5, 2022 2855 Views

Analysis of the Ubiquitination and Phosphorylation of Vangl Proteins
Di Feng [...] Bo Gao
Oct 20, 2022 3416 Views

Isoform-specific, Semi-quantitative Determination of Highly Homologous Protein Levels via CRISPR-Cas9-mediated HiBiT Tagging
Kristina Seiler [...] Mario P. Tschan
Jul 20, 2023 2519 Views
Abstract
Nicotinamide adenine dinucleotide (NAD+) is a coenzyme for many NAD+-consuming proteins with diverse biological functions. Oscillations in NAD+ levels may influence several cellular signaling pathways. NAD+ synthesis via Preiss-Handler route (salvage reactions) has been extensively reported. However, the contribution of L-tryptophan/kynurenine catabolism in de novo NAD+ synthesis is poorly understood. Using L-[14C]-tryptophan tracing in four liver cancer cell lines and siRNA-mediated silencing of arylformamidase (AFMID), a key enzyme involved in L-tryptophan degradation, we demonstrate the contribution of L-tryptophan catabolism in de novo synthesis of NAD+ pools. NAD+ modulation is therefore important in maintaining cellular homeostasis and appropriate cellular functions according to nutrients availability.
Keywords: Kynurenine pathwayMaterials and Reagents
- Liver cancer cell lines: Huh-7 (JCRB, catalog number: JCRB0403 ), HepG2 (ATCC, catalog number: HB-8065 ), SNU-398 (ATCC, catalog number: CRL-2233 ), and SNU-449 (ATCC, catalog number: CRL-2234 )
- DMEM (Life technologies)
- RPMI (Life technologies)
- Amino acid free medium and without fetal bovine serum called Hanks’ Balanced Salt Solution (HBSS) (Sigma-Aldrich)
- Fetal bovine serum (Cultek)
- Penicilin and streptocmycin (Life Technologies, Gibco®)
- 100x Non-essential amino acids’ solution (Life technologies)
- RNAiMAX (Life Technologies, InvitrogenTM, catalog number: 13778030 )
- OptiMem (Life Technologies, catalog number: 11058021 )
- Non silencing small interference RNA control (siCtl) and small interference RNA Arylformamidase (siAFMID) (GE Healthcare Dharmacon, catalog number: L-HUMAN-XX-0005 5 nmol )
- NAD+-[carbonyl-14C] (Perkin Elmer, catalog number: NEC831010UC )
- L-[benzene-ring-U-14C]-tryptophan (Moravek, catalog number: MC 2335 )
- PEI-cellulose thin layer chromatography (TLC) (F plates of 20 x 20 cm) (Merk Millipore catalog number: 105725 )
- Ammonium acetate (Sigma-Aldrich, catalog number: A1542 )
- Ethanol and methanol (Sigma-Aldrich)
- NAD+-[carbonyl-14C] (see Recipes)
- L-[benzene-ring-U-14C]-tryptophan (see Recipes)
- Cell density (around 60-80%) (see Recipes)
Equipment
- Phosphor-image reader (Molecular Dynamics, model: Storm820 )
- Phosphor-image screen (GE Healthcare Dharmacon, catalog number: 63-0034-79 )
- Speedvac centrifuge (Thermo Fisher Scientific)
- 4 °C table top centrifuge (Eppendorf, model: 5424R )
- Fume hood to handle radioactive materials
- Cell scrapers (Corning Incorporated, catalog number: 3008 )
- Ruler
Software
- Fiji software (www.fiji.sc/Fiji)
Procedure
- Four liver cancer cell lines: Huh-7, HepG2, SNU-398 and SNU-449.
- Huh-7, HepG2, SNU-398 and SNU-449 cells were cultured in 10 cm cell culture dishes in presence of complete DMEM (Huh-7 and HepG2 cells) and RPMI medium (SNU-398 and SNU-449). Both media were supplemented with 10% fetal bovine serum and 100 units/ml of penicillin and 0.1 mg/ml of streptomycin.
- Volumes provided are for one well of 12 well plate, and should be scaled according to the size of the well: 200 μl of Optimem is gently mixed with 1.2 μl of either siAFMID or siCtr. This mixture was added to 1.6 μl of RNAiMAX and incubated at room temperature for 20 min in the well.
- Gently resuspend Huh-7, HepG2 (8.0 x 104) cells in 1,000 μl of DMEM or SNU398, SNU449 (3.5 x 104) cells in 1,000 μl of RPMI media and add these cells to the RNAiMAX mixture. The different cell numbers used in this step is to circumvent the variable growth rates in our cell culture conditions to obtain similar cell numbers after the transfection experiment.
- After 24 h, the cell medium was changed to a DMEM or RPMI medium containing 10% fetal bovine serum and 100 units/ml of penicillin and 0.1 mg/ml of streptomycin after removing the supernatant.
- After 4 h, the media was removed, and 1 ml of HBSS medium supplemented with non-essential amino acids (1x) was added to the cell culture.
- Huh-7, HepG2, SNU-398 and SNU-449 cells were cultured in 10 cm cell culture dishes in presence of complete DMEM (Huh-7 and HepG2 cells) and RPMI medium (SNU-398 and SNU-449). Both media were supplemented with 10% fetal bovine serum and 100 units/ml of penicillin and 0.1 mg/ml of streptomycin.
- 2.5 mM of L-[benzene-ring-U-14C]-tryptophan was added to the cells culture in HBSS and in presence of non-essential amino acids. Cells were incubated at 37 °C for 5 h.
- Cell culture medium was removed by discarding the supernatant and, cells were then washed 3 times with ice-cold PBS.
- Lyophilic metabolites were extracted using 200 μl of 80% methanol and 20% water mixture, by scrapping and pipetting vigorously. The mixture was incubated for 10 min at 4 °C.
- Whole cell extracts were then centrifuged at 20,000 x g for 20 min at 4 °C.
- The extracts were concentrated in a speedvac centrifuge to a final volume of 40 μl from 200 μl of 80% methanol and 20% water (step 4) (which might take one hour, you can decrease the time by increasing the vacuum pressure) and subsequently spotted on the PEI-cellulose thin layer chromatography (TLC) F plates.
- Annotating the PEI-cellulose thin layer chromatography (TLC) F plates
- Draw a straight line 1 cm above the bottom of the plate with a pencil.
- Make markings for spotting the metabolic samples equidistant from each other with 2 cm interval as shown in Figure 1.

Figure 1. Preparing the PEI-cellulose thin layer chromatography (TLC) F plate for the assay - 1 μl of NAD+-[carbonyl-14C] was used as a positive control and spotted in the last lane to calibrate the relative migration of labeled metabolites.
- Add 5 μl of metabolic extracts from step 6 and let the spot dry. Repeat this cycle for all the samples until the whole extract is spotted.
- Draw a straight line 1 cm above the bottom of the plate with a pencil.
- Mounting the TLC migration chamber
- Use an apparatus that serves to cast gels for big Western blots as a TLC chamber.
- Wipe the TLC chamber with 70% ethanol.
- Fill the lower tray representing a chamber with migration buffer as described in point-7 of the procedure and as shown in Figure 2.

Figure 2. Mounting the TLC migration chamber
- Use an apparatus that serves to cast gels for big Western blots as a TLC chamber.
- Setup the TLC chamber with the PEI-cellulose TLC plate containing the migration buffer
Figure 3. Setup the TLC chamber with the PEI-cellulose TLC plate containing the migration buffer (composition in step 7)- Carefully mount the TLC plate in which the bottom part of the plate submerses in the migration buffer (composition in step 7). However the spotted samples should not directly be in contact with the migration buffer.
- The plate is mounted vertically to the migration buffer (composition in step 7) and restrained from any movements by sticking it to the top using a tape.
- Carefully mount the TLC plate in which the bottom part of the plate submerses in the migration buffer (composition in step 7). However the spotted samples should not directly be in contact with the migration buffer.
- Annotating the PEI-cellulose thin layer chromatography (TLC) F plates
- Migration buffer composed of 1 M ammonium acetate (pH 5) (30%) and absolute ethanol (70%) used in the bottom reservoir touching 0.5 cm of the PEI-cellulose TLC F plates was used for the mobile phase.
- After 5-6 h of migration reaching about 15 cm from the starting line, the cellulose plate was air-dried and exposed to the radiosensitive phosphor-imager screen for over night. Screen was revealed using Strom-820 phosphor-image reader.
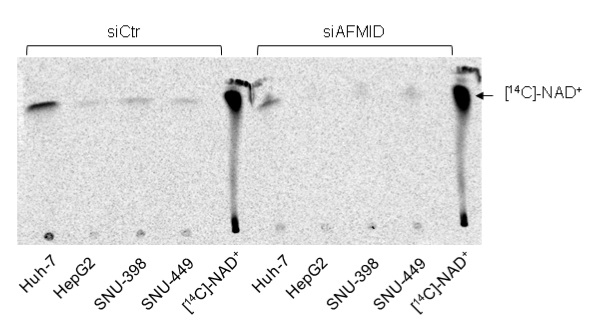
Figure 4. TLC of radiolabelled metabolites derived from L-[14C]-tryptophan in four different liver cancer cell lines. Metabolites from siCtr and siAFMID were loaded simultaneously next to each other. [14C]-NAD+ was used as the calibration control for relative migration of labeled NAD+ derived from L-[14C]-tryptophan (Tummala et al., 2014).
Notes
The experiment has been carried out using the 4 different liver tumor cell lines for at least 4 times and this blot is representative of these 4 different experiments. Consistent incorporation of label has been observed. Fiji software (www.fiji.sc/Fiji) was used to quantify the pixilation of different bands in the image and the relative band intensities were calibrated in respect to the siCtr samples. Data interpretation should be done based on the intensity of the bands at the right size, greater is the band intensity, and greater is the NAD+ content.
Recipes
- NAD+-[carbonyl-14C] used here as a relative migration standard, however it is not mandatory to use this specific isotope, any other phosphor-imager sensitive NAD+ isotope can be used for this purpose.
- L-[benzene-ring-U-14C]-tryptophan has been chosen specifically because of the known contribution of the tryptophan’s benzene ring carbon atom for the NAD+ backbone.
- Cell density (around 60-80%) at the time of starvation yielded most consistent results. The cells are not freshly thawed but maintained in the culture for at least 2-3 passages after thawing.
Note: This method is not validated in other cell lines however, it can be easily adapted to analyze the dependence of different cancer cell lines on de novo NAD+ synthesis.
Acknowledgments
This protocol was adapted as previously reported (Merdanovic et al., 2005). N.D is a recipient of the Spanish Ramón y Cajal fellowship. This work was supported by the Spanish Ministry of Economy and Competitiveness (SAF2010-18518), the Association for International Cancer Research AICR-UK (11-0242), CNIO (BC1104-08) and the European Foundation for the Study of Diabetes (EFSD).
References
- Merdanovic, M., Sauer, E. and Reidl, J. (2005). Coupling of NAD+ biosynthesis and nicotinamide ribosyl transport: characterization of NadR ribonucleotide kinase mutants of Haemophilus influenzae. J Bacteriol 187(13): 4410-4420.
- Tummala, K. S., Gomes, A. L., Yilmaz, M., Grana, O., Bakiri, L., Ruppen, I., Ximenez-Embun, P., Sheshappanavar, V., Rodriguez-Justo, M., Pisano, D. G., Wagner, E. F. and Djouder, N. (2014). Inhibition of de novo NAD(+) synthesis by oncogenic URI causes liver tumorigenesis through DNA damage. Cancer Cell 26(6): 826-839.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tummala, K. S. and Djouder, N. (2015). [14C]-Tryptophan Metabolic Tracing in Liver Cancer Cells. Bio-protocol 5(17): e1582. DOI: 10.21769/BioProtoc.1582.
Category
Cancer Biology > General technique > Biochemical assays > Metabolism
Cell Biology > Cell metabolism > Amino acid
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











