- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Polysomal-mRNA Extraction from Arabidopsis by Sucrose-gradient Separation
Published: Vol 4, Iss 24, Dec 20, 2014 DOI: 10.21769/BioProtoc.1364 Views: 13134
Reviewed by: Tie LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

RNA Purification from the Unicellular Green Alga, Chromochloris zofingiensis
Sean D. Gallaher and Melissa S. Roth
Apr 5, 2018 8150 Views

Extraction of RNA from Recalcitrant Tree Species Paulownia elongata
Niveditha Ramadoss and Chhandak Basu
Jul 20, 2018 7716 Views

Simple Method for Efficient RNA Extraction From Arabidopsis Embryos
Fernanda Marchetti [...] Eduardo Zabaleta
Feb 20, 2025 1913 Views
Abstract
mRNAs surrounded by polysomes are ready for translation into proteins (Warner et al., 1963); these mRNAs are defined as polysomal-mRNAs (Mustroph et al., 2009). The process is affected by various growth conditions or surrounding situations. Microarray analysis is a powerful tool for detecting genome-wide gene expression. Therefore, using polysomal-mRNAs for microarray analysis can reflect the gene translation information (the translatome) under different developmental stages or environmental conditions from eukaryotes. Polysomal-mRNAs can be collected from the polysomal fraction by sucrose-gradient separation for further quantitative PCR or microarray assay. We modified a protocol (Mustroph et al., 2009) for collecting polysomal-mRNAs via sucrose-gradient separation to eliminate monosomal-mRNA contamination from pLAT52:HF:RPL18 Arabidopsis. This transgenic Arabidopsis uses a pollen-specific promoter (ProLAT52) to generate epitope-tagged polysomal-RNA complexes that could be purified with a specific antigen (Lin et al., 2014). The polysomal-mRNAs we obtained via sucrose-gradient separation and antibody purification underwent in vivo translation in pollen tubes grown from self-pollinated gynoecia of Arabidopsis thaliana.
Keywords: Polysomal-mRNAMaterials and Reagents
- pLAT52: HF-RPL18 transgenic Arabidopsis
- RNase-free water
- Tris buffer (Sigma-Aldrich, catalog number: T1378 )
- KCl (Sigma-Aldrich, catalog number: P9541 )
- EGTA (Sigma-Aldrich, catalog number: E3889 )
- MgCl2 (Sigma-Aldrich, catalog number: M8266 )
- β-mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- Cycloheximide (Sigma-Aldrich, catalog number: C7698 )
- Chloramphenicol (Sigma-Aldrich, catalog number: C0378 )
- Polyoxyethylene 10 tridecyl ether (PTE) (Sigma-Aldrich, catalog number: P2393 )
- Sodium deoxycholate (DOC) (Sigma-Aldrich, catalog number: D6750 )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632 )
- Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626 )
- Triton X-100 (Sigma-Aldrich, catalog number: X-100 )
- Polyoxyethylene(23)lauryl ether (Brij -35) (Sigma-Aldrich, catalog number: P6938 )
- Tween-20 (Sigma-Aldrich, catalog number: P1379 )
- NP-40 (Sigma-Aldrich, catalog number: NP 40 )
- Polyoxyethylene (Sigma-Aldrich, catalog number: P2393)
- Deoxycholic acid (Sigma-Aldrich, catalog number: D2510 )
- ANTI-FLAG® M2 Affinity Gel (Sigma-Aldrich, catalog number: A2220 )
- RNAsin (Promega Corporation, catalog number: N2511 )
- 3x FLAG peptide (Sigma-Aldrich, catalog number: F4799 )
- Sucrose (Sigma-Aldrich, catalog number: 84097 )
- Polysomal extraction buffer preparation (see Recipes)
- 20% detergent mixture (see Recipes)
- 20% PTE and 10% DOC (see Recipes)
- Sucrose gradient layer preparation (see Recipes)
- 10x sucrose salts (see Recipes)
- Preparation of the FLAG M2 Agarose beads (see Recipes)
Equipment
- QIAGEN QIA shedder columns (QIAGEN, catalog number: 27115 )
- 15-ml tube (Labcon, catalog number: 9205-946CB-946C )
- Hitachi ultracentrifuge himac CP100WX (Hitachi, catalog number: CP100WX )
- Hitachi P40ST swing rotor (Hitachi, model: P40ST )
- 13PA tubes (Hitachi, catalog number: 332901A )
- Spectrophotometer (Beckman Coulter, catalog number: DU648B )
- UV monitor (GE Healthcare, catalog number: UVIS-920 )
- Pump (GE Healthcare, model: P-50 )
- Fraction collector (Gilson Products, catalog number: FC80 )
- UV detector system (GE Healthcare, catalog number: monitor UVIS-0912 )
- Recorder (GE Healthcare, catalog number: pharmaria LKB REC102 )
Procedure
Pre-treatment: All glass materials were heated in 180 °C for 12 h and all plastic materials were treated with non-sterile DEPC-H2O overnight, then autoclaved at 121 °C for 20 min. RNase-free water was used to prepare all buffers.
- Collect 0.5-1 mg plant tissue sample into a mortar and then add liquid nitrogen immediately.
- After grinding the plant tissue finely, add 1,250 μl polysome extraction buffer into the mortar.
- Transfer the crude extract in an Eppendorf tube and let it sit on ice for 10 min.
- Centrifuge the crude extract for 10 min at 13,000 rpm at 4 °C.
- Transfer the supernatant into a QIA shedder column (700 μl/column). The column is spun for 1 min at 13,000 rpm at 4 °C, then the flow-through is collected.
- Detect the OD260 unit for the flow-through by spectrophotometry.
- Prepare the four different sucrose gradient layers in the Hitachi centrifuge column.
- Carefully add 800 μl sample (20-25 OD260 unit; the nucleic acid concentration for 1 OD260 unit is equal to 40 µg/ml RNA) onto the top sucrose layer (the 20% layer). Balance tubes and buckets to 0.03 g by adding sample or polysome extraction buffer.
- Ultracentrifuge for 225 min at 39,000 rpm at 4 °C in an Hitachi P40AT swing rotor.
- Link the pump with a UV detector and the fraction collector. Link the UV detector with the recorder.
- Put the 260-nm filter into the UV detector system and turn it on at least 20 min before polysome profile analysis.
- Put the probe into a sample separated in sucrose gradient after ultracentrifugation, turn on the pump to suck the sample, and turn on the recorder to create the sucrose gradient profile.
- According to the sucrose gradient profile, combine the polysomal fractions together into a 15-ml tube.
- Add the washed FLAG beads to the 15-ml tube and incubate for 2 h at 4 °C on a rocking platform.
- Centrifuge the mixture for 3 min at 3,000 rpm at 4 °C. Remove the supernatant.
- Add 2 ml wash buffer to the tube and mix gently by inverting it for 5 min at 4 °C, then centrifuge the mixture for 3 min at 3,000 rpm at 4 °C. Remove the supernatant (first wash).
- Add 2 ml wash buffer to the tube and mix gently by inverting it for 5 min at 4 °C, then centrifuge the mixture for 3 min at 3,000 rpm at 4 °C. Remove the supernatant. Repeat 3 times.
- Transfer the mixture to a new Eppendorf tube.
- To elute the affinity-purified polysomes, add the 300 μl FLAG peptide (400 ng/μl) with RNAsin (20 U/ml) to the Eppendorf tube and incubate for 30 min at 4 °C on a rocking platform.
- Centrifuge for 1 min at 8,200 x g at 4 °C. Transfer the supernatant to a new Eppendorf tube. If the supernatant still contains the beads, centrifuge again for 5 min at 13,000 rpm at 4 °C.
- Add additional wash buffer to the eluent to reach 500 μl volume.
- Add phenol/chloroform (1:1) in an equal volume (500 μl) to the eluent and mix by inverting.
- Centrifuge for 10 min at 13,000 rpm at 4 °C.
- Transfer the supernatant to a new Eppendorf tube and add an equal volume of chloroform.
- Centrifuge for 10 min at 13,000 rpm at 4 °C.
- Transfer the supernatant to a new Eppendorf tube and add 0.1 volume 3 M NaOAC (pH 5.2) and 2.5 volume 100% EtOH to precipitate the RNA overnight at -20 °C.
- Centrifuge for 25 min at 13,000 rpm at 4 °C.
- Pour out the supernatant, then wash the pellet with 70% EtOH twice.
- Air-dry the RNA pellet and use 20 μl RNase-free water to suspend the RNA pellet.
Representative data
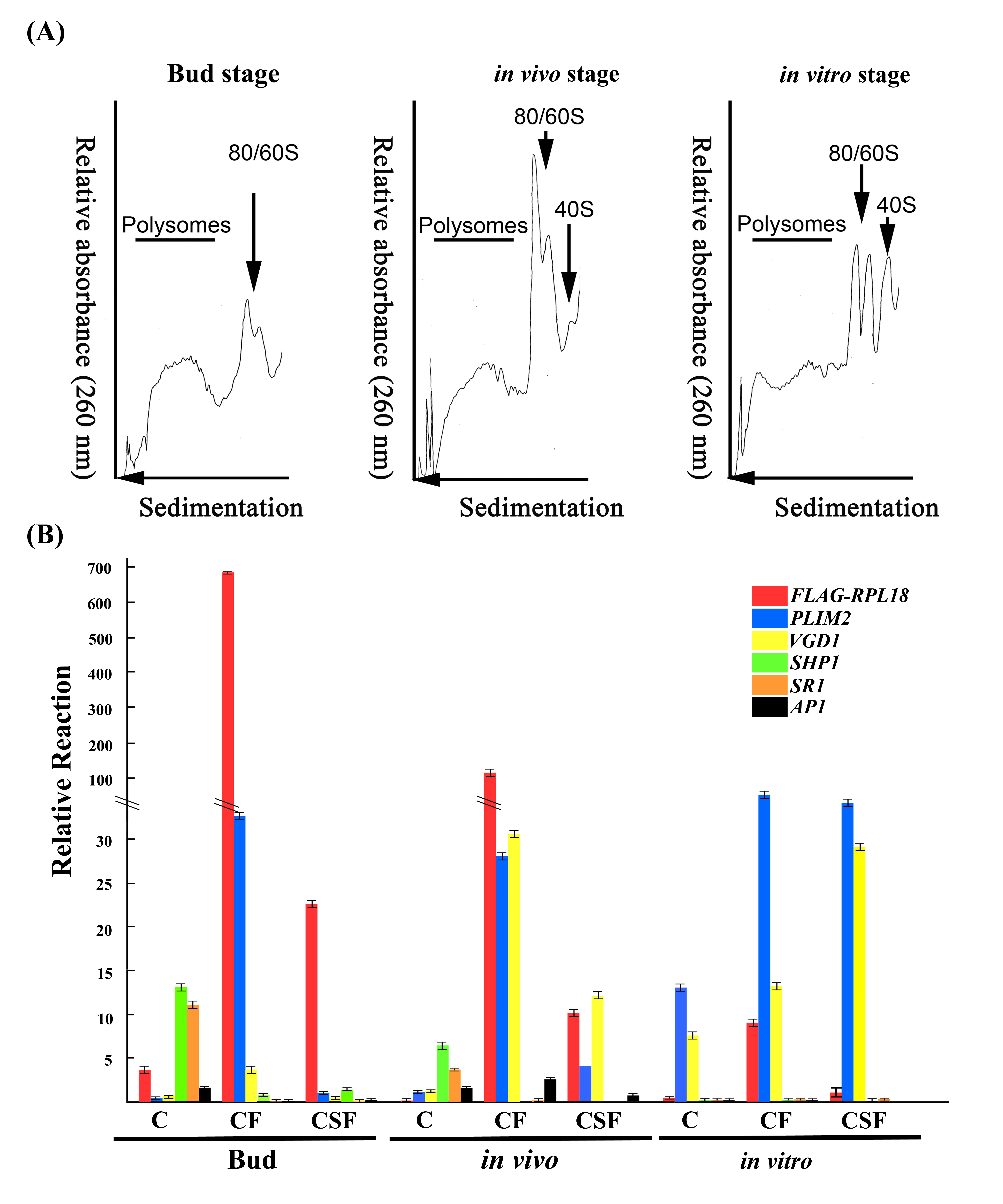
Figure 1. Polysomal profiles and validation of the specificity of immunopurification of mRNAs associated with pollen from pollinated floral buds, in vivo-pollinated pollen tubes, and in vitro-cultured pollen tubes of LAT52:HF-RPL18 transgenic plants. (A) Typical sucrose-gradient absorbance (A 260) profiles of ribosome complexes obtained from Bud stage, in vivo stage, and in vitro-cultured pollen. Positions of peaks corresponding to polysomes (line), 40S ribosomal subunits and 60S ribosomal subunit/80S monosomes (arrowheads) are indicated. The arrow for the sedimentation reflects the sucrose gradient from 20% to 60% (top to bottom). (B) Quantitative RT-PCR with primer sets targeting the FLAG-RPL18 transgene and organ-specific genes to confirm that the RNA extracted from purified polysomal mRNAs examined by sucrose-gradient separation and further purified by FLAG-agarose beads was all male gametophyte-specific. Primers span both the His6-FLAG tag and the RPL18 sequence (FLAG-PRL18); pollen-specific PLIM2 and VGD1, female-specific SHP1 and SR, and petal/sepal-specific AP1. ACT2 was an internal control (Lin et al., 2014).
Table1. Primer pairs for Q-PCR in Figure 1 (Lin et al., 2014)
Notes
- Prepare enough samples for sucrose gradient profile presentation.
Table 2. Number of flowers used for the polysomal-RNA extraction for LAT52:HF-RPL18 Arabidopsis (Lin et al., 2014)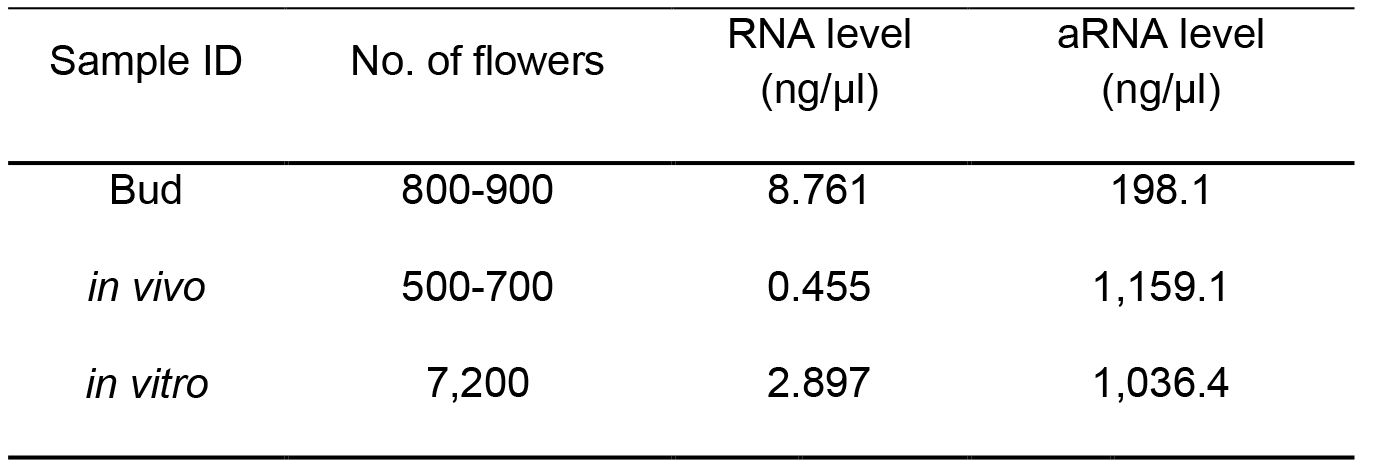
aRNA: antisense RNA synthesized
Recipes
- Polysomal extraction buffer preparation
2 M Tris-HCl (pH 9.0) 1 ml 2 M KCl 1 ml 0.5 M EGTA (pH 8.3) 0.5 ml 1 M MgCl2 0.36 ml dd H2O To 8 ml Mix well β-mercaptoethanol 80 μl 50 mg/ml cycloheximide 10 μl 50 mg/ml chroramphenicol 10 μl 20% detergent mixture 0.5 ml 2% PTE, 10% DOC 1 ml 0.5 M DTT 20 μl 0.5 M PMSF 20 μl ddH2O To 10 ml - 20% detergent mixture
Triton X-100 10 ml Brij 35 10 g Tween-20 10 ml NP-40 10 ml ddH2O To 50 ml - 20% PTE and 10% DOC
Polyoxyethylene 10 trydecyl ether 4 ml Deoxychollic acid 2 g ddH2O To 20 ml - Sucrose-gradient layer preparation
Add 0.1 μl per 1 ml volume of cyclohexamide (50 mg/ml) and chloramphenicol (50 mg/ml) to each layer% sucrose 2 M (68.5%) sucrose 10x sucrose salt ddH2O Vol/gradient 60 44 ml 5 ml 1 ml 1.5 ml 45 66 ml 10 ml 24 ml 3.0 ml 30 44 ml 10 ml 46 ml 3.0 ml 20 14.5 ml 5 ml 30.5 ml 1.5 ml - 10x sucrose salts
Tris-base 2.43 g KCl 0.75 g MgCl2 1.02 g ddH2O To 50 ml Adjust pH value with HCl 8.4 Autoclave 20 min, 121 °C Stored at -20 °C - Preparation of the FLAG M2 Agarose beads
- Use cut pipette to transfer 100 μl FLAG M2 from a bottle into an Eppendorf tube, then centrifuge for 1 min at 8,200 x g at 4 °C.
- Remove the supernatant and add 500 μl wash buffer to resuspend the beads. Centrifuge for 1 min at 8,200 x g at 4 °C.
- Repeat the above step once.
- Wash buffer.
2 M Tris-HCL (pH 9.0) 1 ml 2 M KCl 1 ml 0.5 M EGTA (pH 8.3) 0.5 ml 1 M MgCl2 0.36 ml 50 mg/ml cycloheximide 10 μl 50 mg/ml chroramphenicol 10 μl 0.5 M DTT 20 μl 0.5 M PMSF 20 μl RNAsin 20 U/ml ddH2O To 10 ml
- Use cut pipette to transfer 100 μl FLAG M2 from a bottle into an Eppendorf tube, then centrifuge for 1 min at 8,200 x g at 4 °C.
Acknowledgments
We thank Dr. Julia Bailley-Serres (Department of Botany and Plant Sciences, University of California, Riverside) for the pLAT52-HF-RPL18 Arabidopsis and for sharing the polysomal-mRNA extraction protocol. This work was funded by Academia Sinica (Taiwan), the Taiwan National Science and Technology Program for Agricultural Biotechnology (Lin et al 31; NSTP/AB, 098S0030055-AA) and the Taiwan National Science Council (99-2321-B-001-036-MY3 and 102-2321-B-001-040-MY3) to G.-Y. Jauh.
References
- Lin, S. Y., Chen, P. W., Chuang, M. H., Juntawong, P., Bailey-Serres, J. and Jauh, G. Y. (2014). Profiling of translatomes of in vivo-grown pollen tubes reveals genes with roles in micropylar guidance during pollination in Arabidopsis. Plant Cell 26(2): 602-618.
- Mustroph, A., Juntawong, P. and Bailey-Serres, J. (2009). Isolation of plant polysomal mRNA by differential centrifugation and ribosome immunopurification methods. Methods Mol Biol 553: 109-126.
- Warner, J. R., Knopf, P. M. and Rich, A. (1963). A multiple ribosomal structure in protein synthesis. Proc Natl Acad Sci U S A 49: 122-129.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lin, S. and Jauh, G. (2014). Polysomal-mRNA Extraction from Arabidopsis by Sucrose-gradient Separation . Bio-protocol 4(24): e1364. DOI: 10.21769/BioProtoc.1364.
Category
Plant Science > Plant molecular biology > RNA > RNA extraction
Molecular Biology > RNA > RNA extraction
Biochemistry > RNA > mRNA translation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









