- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
TGFβ Stimulation Assay
Published: Vol 4, Iss 23, Dec 5, 2014 DOI: 10.21769/BioProtoc.1313 Views: 16001
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Imaging Membrane Proteins Using Total Internal Reflection Fluorescence Microscopy (TIRFM) in Mammalian Cells
Kirin D. Gada [...] Leigh D. Plant
Feb 20, 2023 1991 Views

Quantitative Measurement of the Kinase Activity of Wildtype ALPK1 and Disease-Causing ALPK1 Mutants Using Cell-Free Radiometric Phosphorylation Assays
Tom Snelling
Nov 20, 2024 1581 Views

Accurate Identification of Cell Cycle Stages in RPE1 Cells Using the ImmunoCellCycle-ID Method
Syon Reddy [...] Aussie Suzuki
Aug 5, 2025 1870 Views
Abstract
TGFβ is part of a growth factor superfamily which modulates cell growth, differentiation, adhesion, migration, ECM synthesis and apoptosis (Massague, 1998; Siegel and Massague, 2003). Free TGFβ binds to its high affinity TGFβ receptor, a receptor serine/threonine kinase, inducing phosphorylation of Smad2/3 which subsequently forms a complex with Smad4 to translocate to the nucleus where it interacts with multiple co-activators and repressors generating distinct transcriptional responses.
Indeed, TGFβ signaling shows a remarkable cellular context dependency and apparent multifunctionality: e.g. TGFβ is able to inhibit cell proliferation in many epithelial cells but can also enhance proliferation in fibroblasts and cell growth in endothelial cells (Guasch et al., 2007; Xiao et al., 2012); it enhances stem cell pluripotency, but promotes differentiation in other cells (Park, 2011); in cancer development it suppresses pre-malignant cell proliferation, but at the same time promotes conversion to a metastatic phenotype (Chaudhury and Howe, 2009).
The TGFβ stimulation assay monitors the responsiveness of cells to TGFβ. Upon TGFβ stimulation short-term effects such as Smad2 phosphorylation and long-term effects such as cell proliferation can be analyzed. The assay will be described for murine keratinocytes, where TGFβ strongly inhibits cell proliferation, but both assays are applicable for other cell types as well.
Materials and Reagents
- Cell line(s) of interest [here primary murine keratinocytes isolated as described in Montanez et al. (2007)]
- Antibodies
- Bovine serum albumin (BSA) fraction V (Carl Roth, catalog number: 8076 )
- Calcium chloride (CaCl2) (Carl Roth, catalog number: A119.1 )
- Chelex 100 resin (Bio-Rad Laboratories, catalog number: 143-2832 )
- Collagen I (PureCol, Advanced BioMatrix, catalog number: 5005-B )
- Fibronectin (Life Technologies, Gibco®, catalog number: 33016-015 )
- Protein standard (e.g. precision plus protein kaleidoscope standards) (Bio-Rad Laboratories, catalog number: 161-0375 )
- Fetal bovine serum (FBS) (Life Technologies, Gibco®, catalog number: 10270-106 )
- Minimum essential medium (MEM) (Sigma-Aldrich, catalog number: M8167 )
- Insulin (Sigma-Aldrich, catalog number: I5500 )
- Epidermal growth factor (EGF) (Sigma-Aldrich, catalog number: E4127 )
- Transferin (Sigma-Aldrich, catalog number: T8158 )
- Phosphoethanolamine (Sigma-Aldrich, catalog number: P0503 )
- Ethanolamine (Sigma-Aldrich, catalog number: E0135 )
- Hydrocortisone (Calbiochem, catalog number: 386698 )
- Trypsin powder (Life Technologies, Gibco®, catalog number: 27250-018 )
- L-Glutamine (Life Technologies, InvitrogenTM, catalog number: 25030-081 )
- Penicillin-streptomycin (pen-strep) (Life Technologies, InvitrogenTM, catalog number: 15070-063 )
- Click-iT® EdU Alexa Fluor® 488 Imaging Kit (Life Technologies, InvitrogenTM, catalog number: C10420 )
- Nonfat dried milk powder (AppliChem, catalog number: A0830 )
- PVDF membrane (e.g. Immobilon-P) (Merck KGaA, catalog number: IPVH00010 )
- Recombinant TGFβ1 (PeproTech, catalog number: 100-21 )
- Tween20 (Sigma-Aldrich, catalog number: P9416 )
- Sodium dodecyl sulfate (SDS) (Carl Roth, catalog number: 0183.1 )
- β-Mercaptoethanol (Carl Roth, catalog number: 4227.3 )
- Bromophenol blue (Carl Roth, cataog number: A512.1 )
- Sodium chloride (NaCl) (Carl Roth, catalog number: P029 )
- Sodium hydrogen carbonate (NHCO3) (Carl Roth, catalog number: 6885 )
- Sodium carbonate (Na2CO3) (Carl Roth, catalog number: A135 )
- Sodium deoxycholate (Sigma-Aldrich, catalog number: D6750 )
- di-Sodium hydrogen phosphate (Na2HPO4) (Carl Roth, catalog number: T876 )
- Glycine (Carl Roth, catalog number: 0079 )
- Glycerol (Carl Roth, catalog number: 4043.1 )
- Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 258148 )
- Methanol (Sigma-Aldrich, catalog number: 322415 )
- Potassium chloride (KCl) (Carl Roth, catalog number: HN02.3 )
- Potassium dihydrogen phosphate (KH2PO4) (Carl Roth, catalog number: 3904 )
- Nonidet-P40 (NP-40) (e.g. IGEPAL CA-630) (Sigma-Aldrich, catalog number: I8896 )
- Protease inhibitor cocktail (e.g. cOmplete) (Roche, catalog number: 04693116001 )
- Phosphatase inhibitor cocktail 2 (Sigma-Aldrich, catalog number: P5726 )
- Phosphatase inhibitor cocktail 3 (Sigma-Aldrich, catalog number: P0044 )
- 2-Propanol (Carl Roth, catalog number: T902.1 )
- Tris-HCl (Carl Roth, catalog number: 9090.3 )
- Cell type specific growth (see Recipes)
- Cell type specific starving medium (see Recipes)
- Phosphate-buffered saline (PBS) (see Recipes)
- Cell lysis buffer supplemented with protease and phosphatase inhibitors (see Recipes)
- Standard SDS-PAGE running buffer (see Recipes)
- 2x Laemmli sample buffer (see Recipes)
- Western blot transfer buffer (see Recipes)
- TBS (see Recipes)
- TBS-T (see Recipes)
- Western blot stripping buffer (see Recipes)
Equipment
- 6-well plate and 10 cm dishes for cell culture
- Standard centrifuge to spin down cells and protein lysis
- Cell scraper (e.g. Corning cell scraper) (Sigma-Aldrich, catalog number: CLS3010 )
- Standard SDS-PAGE and Western blot equipment
- Cell culture Incubator 37 °C, 5% CO2 for standard cell culture conditions
- Flow cytometer (e.g. BD FACS Canto) using FlowJo as analysis software
Procedure
- Analysis of proliferation after TGFβ stimulation (long-term effect)
Note: Each condition should be measured in technical triplicates. For keratinocytes one well of a 6 well plate is sufficient for FACS analysis but depending on the cell type, the assay can be performed with higher or lower amounts of cells. Proliferation is measured by uptake of the nucleotide analogue 5-ethynyl-2′-deoxyuridine (EdU) during cell cycle S-Phase and then detected by Click-iT chemistry. 5-brom-2’-deoxyuridine (BrdU) uptake can be used alternatively.
- Plate cell line of interest in complete medium in such a density, so that wells will reach a confluency of 70% the next day (1.2 x 105 cells/well for keratinocytes). For keratinocytes wells should be coated with coating medium for 30 min at 37 °C which should be removed before plating the cells).
- The next day medium is removed and (without any washing step) is replaced with cell culture medium (should be room temperature) supplemented with reduced serum concentration (here 5% FBS) and 0, 5 or 10 ng/ml TGFβ1 and continue culture under standard conditions (37 °C, 5% CO2).
- After 6 h add 10 µM EdU per well without changing the medium and continue to culture cells under standard conditions (37 °C, 5% CO2).
- Further incubate the cells for 8 h and then generate single cell suspension by trypsin digestion. For trypsin digestion remove the medium and wash cells once with PBS. Then add trypsin solution, incubate cells at standard condition (37 °C, 5% CO2) and as soon as cells have detached collect cells by adding complete medium. After spinning down the cells (5 min at 900 rpm, 78 x g) wash single cell suspension once with PBS before following Click-iT assay kit protocol.
Note: The isolation time point can vary between cell types. It is recommended to determine in a pre-experiment a time point, where approximately 70% of the untreated cells of interest are EdU positive.
- Click-iT protocol (mainly follows manufacturers protocol which can be downloaded on the Invitrogen web site).
- Wash cells once with 1% BSA-PBS solution at room temperature.
Note: Cell washes and buffer changes are performed by centrifugation of cell suspensions for 2 min at 2,100 rpm (380 x g), removal of the supernatant and gentle resuspension of the cells in the respective new buffer.
- Fix cells with 200 µl 3.7% formaldehyde in PBS (Component D in the kit) for 15 min at room temperature and permeabilize by buffer change to 200 µl 1x Click-iT saponin-based permeabilization solution (Component E in the kit) and incubate for 15 min.
- Prepare Click-iT staining solution according manufacturer protocol using PBS (see Click-iT reaction cocktail table) and incubate cells with this solution for 30 min in the dark at room temperature.
- Wash cells once with 500 µl Click-iT permeabilization and washing buffer, resuspend cells in 200 µl of the same buffer and immediately proceed to FACS measurement.
- To detect Alexa Fluor 488 for EdU labeling, use 488 nm excitation with a green emission filter 530/30 nm or similar (FL1 channel).
- Use unstained cells as negative control.
- Determine percentage of EdU positive cells, identifying all cells which have been in cell cycle S-phase during EdU incorporation (=proliferating cells).
- Wash cells once with 1% BSA-PBS solution at room temperature.
- Plate cell line of interest in complete medium in such a density, so that wells will reach a confluency of 70% the next day (1.2 x 105 cells/well for keratinocytes). For keratinocytes wells should be coated with coating medium for 30 min at 37 °C which should be removed before plating the cells).
- pSmad detection after TGFβ stimulation (short-term effect, or eminent effects)
Note: Phosphorylation of Smad proteins is an immediate effect after TGFβ receptor activation of the canonical TGFβ signaling cascade and will occur in all cell types. Conversely the subsequent TGFβ downstream signaling can be highly diverse between different cell types.
- Plate for each condition one 10 cm dish. Cell of interest density should be chosen so that 10 cm dishes will reach a confluency of 70% the next day (for keratinocytes 2 x 106 cells/dish). For keratinocytes dishes should be coated with coating medium for 30 min at 37 °C which should be removed before plating the cells.
- The next day serum-starve the cells for several hours (2-4 h) in starving medium (e.g. Starving KGM) under standard cell culture conditions.
- Then add 5 ng/ml TGFβ1 to the starving medium and isolate cells after 0, 30, 60 and 120 min continued incubation at standard cell culture conditions.
- For cell isolation, wash cells once with PBS and lyse them in appropriate volume of lysis buffer (e.g. RIPA, for keratinocytes 120 µl per 10 cm dish) directly on the plate on ice. Collect cell lysates with a cell scraper and pipette up and down 5 times with a 200 µl tip for complete cell lysis. Then spin down cell lysates at full speed (10 min, 14,000 rpm, 16,800 x g) at 4 °C and isolate the supernatant (sample to run on SDS-page).
- Separate samples on a 10% SDS-PAGE using standard Western blot protocol considering that Smad2 migrates around 55-60 kDa. A lane with pre-stained protein standard (e.g. precision plus protein kaleidoscope standards) should be included (for keratinocytes, 20 µg of protein per lane was ideal.) After gel separation, samples were transferred onto a PVDF membrane, using over-night tank transfer and Dunn buffer. Recipes for 2x Laemmli sample buffer, standard SDS-PAGE running buffer, Dunn Carbonate buffer are included in the recipes section.
- For the detection of murine pSmad2 and total Smad2/3 the following antibodies and conditions were used:
- pSmad2:
- Block membrane for 1 h at room temperature with 5% milk TBS-T and incubate pSmad2 antibody 1:1,000 in 1% milk TBS-T over night at 4 °C.
- Wash membranes 3x 15 min with TBS-T and incubate with secondary antibody for 1 h at room temperature.
- Wash membranes 3x 15 min with TBS-T before detection.
- Block membrane for 1 h at room temperature with 5% milk TBS-T and incubate pSmad2 antibody 1:1,000 in 1% milk TBS-T over night at 4 °C.
- Total Smad2/3:
- Block membrane for 1 h at room temperature with 5% BSA TBS-T and incubate total Smad2/3 antibody 1:1,000 in 1% BSA TBS-T over night at 4 °C.
- Wash membranes 3x 15 min and incubate with secondary antibody for 1 h at room temperature.
- Wash membranes 3x 15 min before detection.
Note: It is recommended to detect first pSmad2 and after membrane stripping to blot for total Smad2/3.
For membrane stripping incubate membrane for 10 min in pre-heated (50 °C) stripping buffer at 50 °C. Then wash the membrane with ddH2O (4x 5 min) before reactivating the membrane with 2-propanol. Repeat the washing of the membranes (4x 5 min) before blocking.
- Block membrane for 1 h at room temperature with 5% BSA TBS-T and incubate total Smad2/3 antibody 1:1,000 in 1% BSA TBS-T over night at 4 °C.
- pSmad2:
- Plate for each condition one 10 cm dish. Cell of interest density should be chosen so that 10 cm dishes will reach a confluency of 70% the next day (for keratinocytes 2 x 106 cells/dish). For keratinocytes dishes should be coated with coating medium for 30 min at 37 °C which should be removed before plating the cells.
Representative data
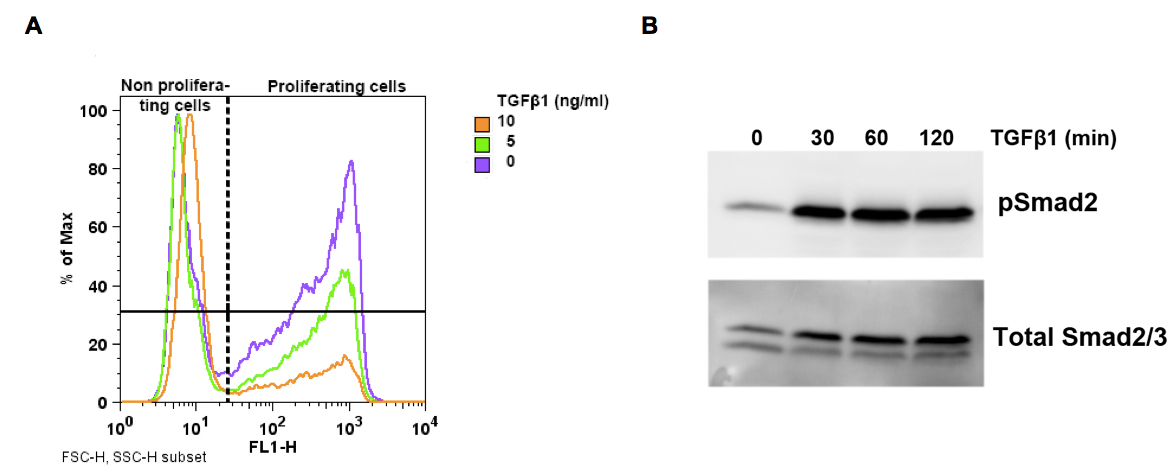
Figure 1. A. Representative FACS histogram of murine kerationcytes using Click-iT EdU Alexa Fluor 488 assay kit. Keratinocytes were incubated for 6 h with indicated TGFβ1 concentrations before addition of 10 µM EdU. Cells were analyzed after 8 h of EdU incorporation to detect proliferating cells. Note the decrease of proliferating cells with increasing TGFβ1 concentration. B. Representative Western bot for pSmad2 detection in murine kerationcytes after stimulation with 5 ng/ml TGFβ1 for indicated time points.
Recipes
- Cell type specific growth and starving medium as well as trypsin solution (shown for murine keratinocyte culture)
- Keratinocyte growth medium (KGM) Filter the mixture through 0.2 µm and stored at 4 °C for up to 1 month.
Final working concentration
Initial stock-concentration
Vol.
MEM
500 ml
5 µg/ml insulin
5 mg/ml in 4mM HCl
0.5 ml
10 ng/ml EGF
200 µg/ml in PBS
25 µl
10 µg/ml transferin
5 mg/ml in PBS
1 ml
10 µM phosphoethanolamine
10 mM in PBS
0.5 ml
10 µM ethanolamine
10 mM in PBS
0.5 ml
0.36 µg/ml hydrocortisone
5 mg/ml in ethanol
36 µl
1x glutamine
100x
5 ml
1x pen-strep
100x
5 ml
8% chelated FBS (Ca2+ free) (see Recipes)
40 ml
45 µM CaCl2 (sterile filtrated)
100 mM
225 µl
- Starving KGM
Filter the mixture through 0.2 µm and store at 4 °C for up to 1 monthFinal working concentration
Initial Stock-concentration
Vol.
MEM
500 ml
1x pen-strep
100x
5 ml
45 µM CaCl2 (sterile filtrated)
100 mM
225 µl
- Keratinocyte trypsin (0.4%)
Dissolve 0.4 g trypsin powder in 100 ml PBS and pass through a 0.2 µm filter for sterilization. It can be stored either at -20 °C for 1 year or at 4 °C for 1 month
Avoid repeated freeze thaw cycles
- Chelated FBS
Add hydrated Chelex 100 resin to FBS (20 g of resin for 40 ml of FBS) and stir for 1 h at 4 °C
Further remove the resin by filtration and store chelated FBS at -20 °C
- Coating medium for keratinocytes
Dilute Collagen I (30 µg/ml) and Fibronectin (10 µg/ml) in PBS directly before use
- Keratinocyte growth medium (KGM)
- Phosphate-buffered saline (PBS)
Dissolve NaCl (137 mM) 8 g/L, KCl (2.7 mM) 0.2 g/L, Na2HPO4 (10 mM) 1.44 g/L, KH2PO4 (1.8 mM) 0.24 g/L in ddH2O and adjust pH to 7.4 with HCl
- Cell lysis buffer supplemented with protease and phosphatase inhibitors (for kerationcytes RIPA)
For RIPA buffer dissolve Tris-HCl (50 mM, pH 7.4), NP-40 (1%), Na-deoxycholate (0.5%), SDS (0.1%), NaCl (150 mM) and EDTA (2 mM)
(Prior to use add protease inhibitors and phosphatase inhibitor 2 and 3)
- Standard SDS-PAGE running buffer
To prepare 10x running buffer dissolve Tis-HCl 30 g/L, Glycine 144 g/L and 10 g/L SDS in ddH2O
Dilute to 1x before use
- 2x Laemmli sample buffer
Dissolve Tris-HCl (100 mM, pH 6.8), 20% glycerol, 2% SDS and 0.01% bromophenol blue in ddH2O
Add β-mercaptoethanol (4%) just before use
- Western blot transfer buffer (e.g. Dunn Carbonate buffer)
Here Dunn Carbonate buffer is used
For 1x Dunn Carbonate Buffer dissolve NaHCO3 (10mM) 0.84 g/L, Na2CO3 (3 mM) 0.318 g/L and methanol (20%) 200 ml in ddH2O
- TBS
Dissolve Tris-HCl (50 mM) 6.05 g/L and NaCl (150 mM) 8.76 g/L in ddH2O and adjust pH to 7.5 with HCl
- TBS-T
TBS supplemented with 0.1% Tween20
- Western blot stripping buffer
2% SDS
62.5 mM Tris-HCl (pH 6.7)
100 mM β-Mercaptoethanol (add only directly before use)
Acknowledgments
This work was funded by the Max Planck Society.
References
- Chaudhury, A. and Howe, P. H. (2009). The tale of transforming growth factor-beta (TGFbeta) signaling: a soigne enigma. IUBMB Life 61(10): 929-939.
- Guasch, G., Schober, M., Pasolli, H. A., Conn, E. B., Polak, L. and Fuchs, E. (2007). Loss of TGFbeta signaling destabilizes homeostasis and promotes squamous cell carcinomas in stratified epithelia. Cancer Cell 12(4): 313-327.
- Massague, J. (1998). TGF-beta signal transduction. Annu Rev Biochem 67: 753-791.
- Montanez, E., Piwko-Czuchra, A., Bauer, M., Li, S., Yurchenco, P. and Fassler, R. (2007). Analysis of integrin functions in peri-implantation embryos, hematopoietic system, and skin. Methods Enzymol 426: 239-289.
- Park, K. S. (2011). Tgf-Beta family signaling in embryonic stem cells. Int J Stem Cells 4(1): 18-23.
- Siegel, P. M. and Massague, J. (2003). Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer 3(11): 807-821.
- Xiao, L., Du, Y., Shen, Y., He, Y., Zhao, H. and Li, Z. (2012). TGF-beta 1 induced fibroblast proliferation is mediated by the FGF-2/ERK pathway. Front Biosci (Landmark Ed) 17: 2667-2674.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rognoni, E. (2014). TGFβ Stimulation Assay. Bio-protocol 4(23): e1313. DOI: 10.21769/BioProtoc.1313.
Category
Cell Biology > Cell signaling > Phosphorylation
Cell Biology > Cell viability > Cell proliferation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










