- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Detection of S-acylation on Recombinant Proteins via the Biotin-Switch Technique
Published: Vol 4, Iss 22, Nov 20, 2014 DOI: 10.21769/BioProtoc.1296 Views: 10202
Reviewed by: Zhaohui LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Detection of Disulfides in Protein Extracts of Arabidopsis thaliana Using Monobromobimane (mBB)
Shin‐nosuke Hashida and Maki Kawai-Yamada
Mar 5, 2019 6399 Views

Simple Method to Determine Protein Redox State in Arabidopsis thaliana
Keisuke Yoshida and Toru Hisabori
Jun 5, 2019 7443 Views

In vitro Auto- and Substrate-Ubiquitination Assays
Hye Lin Park [...] Gyeong Mee Yoon
Apr 5, 2022 3516 Views
Abstract
Protein palmitoylation is the post-translational modification of proteins via the attachment of palmitate through acyl linkages. The nucleophile sulfhydryl group of cysteines is the common palmitoylation site. Covalent attachment of palmitate occurs on numerous proteins and is usually associated with directing protein localization to the endomembrane system. Detection of protein palmitoylation by in vivo labeling with tritium-labeled palmitic acid typically requires an autoradiographic exposure time of several months, and, thus is not suitable for rapid analyses. Here, we described an easy protocol for quick in vitro detection of protein S-acylation using the Arabidopsis protein kinase, PBS1, as an example. To determine whether PBS1 is modified through thioester linkage to acyl groups, we employed a “biotin switch” assay (Hemsley et al., 2008). This work was first published in Qi et al. (2014), but we expand on the method here. PBS1 functions within the basal immune system of plants, and is a target of the bacterial cysteine protease, AvrPphB (Shao et al., 2002; Zhang et al., 2010). It contains a predicted N-terminal S-acylation motif (MGCFSCFDS), with both Cys-3 and Cys-6 residues predicted to be palmitoylated by CSS-Palm 3.0 (http://csspalm.biocuckoo.org/; Ren et al., 2008). Our method utilizes hydroxylamine-induced cleavage of thioester bonds, which results in free sulfhydryl groups that can then be conjugated to a biotin derivative, 1-biotinamido-4-[4′-(maleimidomethyl) cyclohexanecarboxamido]-butane (Biotin-BMCC). The conjugates are detectable by Western blot with streptavidin-horseradish peroxidase. The whole process of in vitro labelling and detection took less than 3 days, allowing the fast detection of protein modifications via thioester bonds such as palmitoylation.
Materials and Reagents
- Nicotiana benthamiana (N. benthamiana) plants
- Agrobacterium tumefaciens strain GV3101 (pMP90)
- Bacto yeast extract (BD Biosciences, catalog number: 288620 )
- Bacto tryptone (BD Biosciences, catalog number: 211699 )
- Magnesium chloride hexahydrate (EMD Millipore, catalog number: 442611 )
- Acetosyringone (Sigma-Aldrich, catalog number: D134406-5G )
- Dexamethasone (Sigma-Aldrich, D4902-1G )
- Trizma Tris base (Sigma-Aldrich, catalog number: 93362 )
- Sodium chloride (EMD Millipore, catalog number: 567442 )
- Nonidet P-40(Sigma-Aldrich, catalog number: 21-3277 )
- Plant proteinase inhibitor cocktail (Sigma-Aldrich, catalog number: P9599-5M )
- SDS (Sigma-Aldrich, catalog number: L3771-100G )
- Glycerol (EMD Millipore, catalog number: 356350 )
- β-mercaptoethanol (Sigma-Aldrich, catalog number: M3148-25ML )
- EDTA (Sigma-Aldrich, catalog number: E6758-100G )
- BSA (Sigma-Aldrich, catalog number: A4503-100G )
- Bromphenol blue (EMD Millipore, catalog number: BX1410 )
- Anti-HA monoclonal antibody matrix (Roche Diagnostics, catalog number: 11867423001 )
- Tris-HEPES-SDS polyacrylamide gels (Thermo Fisher Scientific, catalog number: 25204 )
- Anti-HA peroxidase (Sigma-Aldrich, catalog number: H6533-1VL )
- High Sensitivity Streptavidin-HRP Conjugate (Thermo Fisher Scientific, catalog number: 21130 )
- ImmunoStar HRP Substrate Kit (Bio-Rad Laboratories, catalog number: 170-5070 )
- N-ethylmaleimide (Sigma-Aldrich, catalog number: E3876-5G )
- Hydroxylamine (Sigma-Aldrich, catalog number: 467804-10ML )
- 1-biotinamido-4-[4′(maleimidomethyl)-cyclohexane-carboxamido]-butane (EZ-link BMCC-Biotin) (Thermo Fisher Scientific, catalog number: 21900 )
- Nitrocellulose Membrane (Thermo Fisher Scientific, catalog number: WP4HY00010 )
- Metro-Mix 360 (Sun Gro Horticulture Canada)
- LB liquid medium (see Recipes)
- Lysis buffer (see Recipes)
- 4x SDS loading buffer (see Recipes)
- 5x nonreducing protein sample buffer (see Recipes)
Equipment
- Confocal microscope system (Leica Microsystem, model: TCS SP5 )
- Mini-Protean Electrophoresis and Blotting system (Bio-Rad Laboratories)
- Benchtop Centrifuge 5424 (Eppendorf)
- Lab tube rotator (Thermo Fisher Scientific)
- Gene Pulser (Bio-Rad Laboratories, catalog number: 1652076 )
- 1 ml syringe (BD Biosciences, catalog number: 305945 )
Procedure
- Transient expression assays in N. benthamiana
Transient expression assays were performed as described previously (DeYoung et al., 2012). Briefly:
- N. benthamiana plants were grown in small plastic pots filled with Metro-Mix 360 under an 8/16 h photoperiod at 150 μmol/m/s and 23 °C.
- Dexamethasone-inducible constructs of N20sYFP:HA and n20sYFP:HA were mobilized into Agrobacterium tumefaciens strain GV3101 (pMP90) by electroporation.
- After overnight culture in Luria-Bertani liquid medium, cells (7 ml culture) were pelleted and resuspended in 10 mM MgCl2 solution with 100 μm acetosyringone and suspensions were diluted to OD600 = 0.3 before infiltration.
- The cells were incubated for 2 h at room temperature and infiltrated with a needleless 1 ml syringe into leaves of 4-week-old (best age for transient expression) N. benthamiana plants. A minimum of six fully expanded leaves should be infiltrated for one experiment.
- Transgenes were induced by spraying leaves with 50 µM dexamethasone 40 h after inoculation. Samples were harvested for protein extraction 4 to 6 h after dexamethasone application.
- Laser scanning confocal microscopy was performed at 5 h after dexamethasone application to confirm the expression of sYFP fusions. A 1 cm x 1 cm tissue was clipped from the infiltrated leaf area and placed in a drop of ddH2O in the center of a slide. After a cover slip was placed, the slide was examined using the Leica SP5 system.
- N. benthamiana plants were grown in small plastic pots filled with Metro-Mix 360 under an 8/16 h photoperiod at 150 μmol/m/s and 23 °C.
- Immunoprecipitation and Immunoblotting
- For total protein extraction, six infiltrated leaves were ground in lysis buffer. Tissues can be flash-frozen in liquid nitrogen and stored at -80 °C till needed.
- Homogenates were centrifuged twice at 13,000 rpm at 4 °C for 10 min, and supernatants were transferred to new tubes.
- 100 μl an anti-HA monoclonal antibody matrix was added into each homogenate. The mixture was then incubated at 4 °C overnight and rotated end-to-end.
- The resin was pelleted by centrifuging at 16,000 x g for 10 sec and washed three times with 1 ml lysis buffer.
- For immuno-blot, the immuno-precipitates were mixed with 4x SDS loading buffer at a ratio of 3:1 and boiled for 10 min before resolving on 4% to 20% (w/v) gradient Tris-HEPES-SDS PAGE.
- Protein samples were then transferred to a nitrocellulose membrane for probing with anti-HA peroxidase.
- The ImmunoStar HRP Substrate Kit was used for detecting antibody complexes.
- For total protein extraction, six infiltrated leaves were ground in lysis buffer. Tissues can be flash-frozen in liquid nitrogen and stored at -80 °C till needed.
- Detection of S-Acylation
The in vitro detection of PBS1 S-acylation was performed as described by Berzat et al. (2005). Briefly:
- The immuno-precipitates of N20-sYFP:HA or n20-sYFP:HA were incubated in 1 ml lysis buffer with 50 mM N-ethylmaleimide on an end-to-end rotator for 48 h at 4 °C to block free sulfhydryl groups.
- The immuno-precipitates were then washed, incubated in 500 μl 1 M hydroxylamine solution on an end-to-end rotator for 1 h at room temperature to hydrolyze Cys-palmitate thioester bonds.
- The immuno-precipitates were washed and incubated in 500 μl 50 mM Tris (pH 7.0) solution containing 1 μm EZ-Link Biotin-BMCC on an end-to-end rotator for 2 h at room temperature to label cleaved thioester bonds.
- The immuno-precipitates were washed, resuspended in nonreducing protein sample buffer, resolved on 4% to 20% (w/v) gradient Tris-HEPES-SDS PAGE.
- The protein samples were transferred to a nitrocellulose membrane.
- The membrane was blocked in 5% biotin-free BSA TBST solution for 1 h and then probed with streptavidin-horseradish peroxidase.
- The ImmunoStar HRP Substrate Kit was used for detecting biotin-streptavidin complexes.
- The immuno-precipitates of N20-sYFP:HA or n20-sYFP:HA were incubated in 1 ml lysis buffer with 50 mM N-ethylmaleimide on an end-to-end rotator for 48 h at 4 °C to block free sulfhydryl groups.
Representative data
Representative data are shown in Figure 1. This experiment was repeated three times using this protocol with similar results.
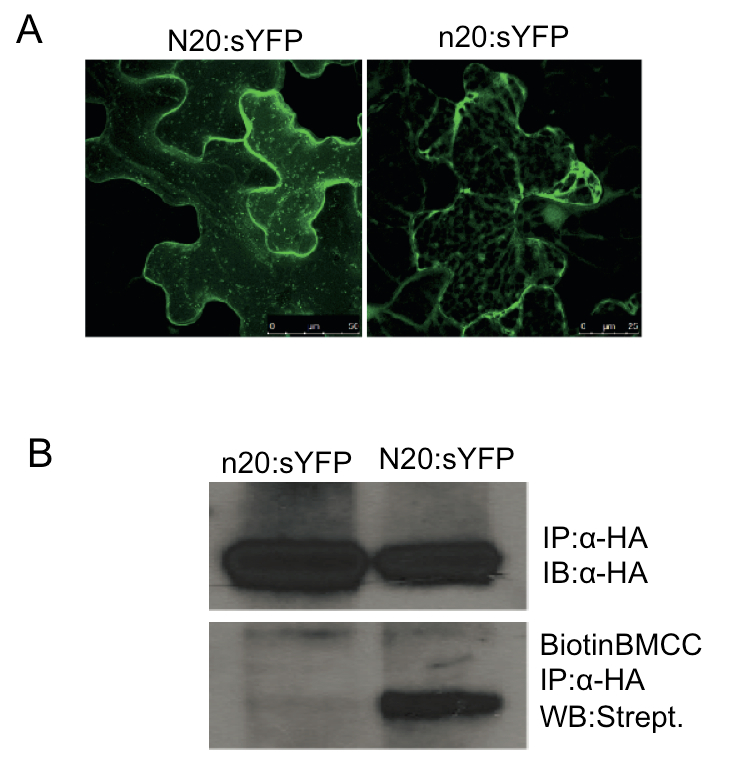
Figure 1. S-Acylation mediates PBS1 localization to the PM. A. The predicted N-terminal S-acylation motif in PBS1 is required for plasma membrane localization. N20:sYFP indicates fusion of the first 20 amino acids of PBS1, which contains the predicted palmitoylation motif, to the super yellow fluorescent protein. n20 is the fragment with the G2A/C3AC/6A triple mutations. Both fusion proteins were transiently expressed in Nicotiana benthamiana. Confocal microscopy was performed at 5 h post dexamethasone induction. All images are three-dimensional projections from a Z-stack. The N20:sYFP localized to the plasma membrane and mobile vesicle-like structures were observed, which was reported when acylated proteins were overexpressed (Vilas et al., 2006; Joensuu et al., 2010). However, the n20:sYFP fusion was detected exclusively as cytoplasmic strands. The YFP signal was showed in the false color of green. B. PBS1 is S-acylated. Cell extracts of N. benthamiana tissue expressing N20:sYFP-HA or n20:sYFP-HA were subjected to immunoprecipitation using anti-HA matrix (IP). Immunoprecipitates were subjected to immuno-blot with anti-HA antibodies (IB) as the loading control. At the same time, the immunoprecipitates were treated with 50 mM N-ethylmaleimide to block free sulfhydryl groups, incubated with 1 M hydroxylamine to hydrolyze any Cys-palmitate thioester bonds, and then treated with 1 μm EZ-Link Biotin-BMCC in 50 mM Tris (pH 7.0) to label the newly exposed free sulfhydryl groups resulting from cleaved thioester bonds. The modified immunoprecipitates were resolved by SDS-PAGE and analyzed by Western blot (WB) with streptavidin horseradish peroxidase. Reproduced from Qi et al. (2014) (Copyright American Society of Plant Biologists).
Notes
- No strong reductants, such as DTT or β-mercaptoethanol, can be used in any step of the sample preparation.
- Confocal microscopy was employed merely for quick confirmation of protein expression. It is not necessary for the following immuno-precipitation and in vitro labelling.
- This technique can be extended to recombinant proteins expressed in other eukaryotic heterologous expression systems such as Pichia pastoris, Spodoptera frugiperda, and fibroblast, etc.
Recipes
- LB liquid medium (per liter)
10 g tryptone
5 g yeast extract
10 g NaCl
- Lysis buffer
50 mM Tris (pH 7.5)
150 mM NaCl
0.1% (v/v) Nonidet P-40
1% (v/v) plant proteinase inhibitor cocktail
- 4x SDS loading buffer
50 mM Tris-HCl (pH 6.8)
2% (w/v) SDS
10% (v/v) glycerol
1% (v/v) β-mercaptoethanol
12.5 mM EDTA
0.02% (w/v) bromphenol blue
- 5x nonreducing protein sample buffer
10% (w/v) SDS
1 M Tris-HCl (pH 6.8)
25% (w/v) sucrose
0.01% (w/v) bromphenol blue
Acknowledgments
This work was supported by the National Institute of General Medical Sciences at the National Institutes of Health (grant no. R01 GM046451 to R.W.I). This protocol was adapted from a previous work by Qi et al. (2014).
References
- Ade, J., DeYoung, B. J., Golstein, C. and Innes, R. W. (2007). Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A 104(7): 2531-2536.
- Berzat, A. C., Buss, J. E., Chenette, E. J., Weinbaum, C. A., Shutes, A., Der, C. J., Minden, A. and Cox, A. D. (2005). Transforming activity of the Rho family GTPase, Wrch-1, a Wnt-regulated Cdc42 homolog, is dependent on a novel carboxyl-terminal palmitoylation motif. J Biol Chem 280(38): 33055-33065.
- DeYoung, B. J., Qi, D., Kim, S. H., Burke, T. P. and Innes, R. W. (2012). Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol 14(7): 1071-1084.
- Hemsley, P. A., Taylor, L. and Grierson, C. S. (2008). Assaying protein palmitoylation in plants. Plant Methods 4: 2.
- Joensuu, J. J., Conley, A. J., Lienemann, M., Brandle, J. E., Linder, M. B. and Menassa, R. (2010). Hydrophobin fusions for high-level transient protein expression and purification in Nicotiana benthamiana. Plant Physiol 152(2): 622-633.
- Qi, D., Dubiella, U., Kim, S. H., Sloss, D. I., Dowen, R. H., Dixon, J. E. and Innes, R. W. (2014). Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 is dependent on S-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1. Plant Physiol 164(1): 340-351.
- Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y. and Yao, X. (2008). CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel 21(11): 639-644.
- Shao, F., Merritt, P. M., Bao, Z., Innes, R. W. and Dixon, J. E. (2002). A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109(5): 575-588.
- Vilas, G. L., Corvi, M. M., Plummer, G. J., Seime, A. M., Lambkin, G. R. and Berthiaume, L. G. (2006). Posttranslational myristoylation of caspase-activated p21-activated protein kinase 2 (PAK2) potentiates late apoptotic events. Proc Natl Acad Sci U S A 103(17): 6542-6547.
- Zhang, J., Li, W., Xiang, T., Liu, Z., Laluk, K., Ding, X., Zou, Y., Gao, M., Zhang, X., Chen, S., Mengiste, T., Zhang, Y. and Zhou, J. M. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7(4): 290-301.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Qi, D. and Innes, R. W. (2014). In vitro Detection of S-acylation on Recombinant Proteins via the Biotin-Switch Technique. Bio-protocol 4(22): e1296. DOI: 10.21769/BioProtoc.1296.
-
Qi, D., Dubiella, U., Kim, S. H., Sloss, D. I., Dowen, R. H., Dixon, J. E. and Innes, R. W. (2014). Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 is dependent on S-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE1. Plant Physiol 164(1): 340-351.
Category
Plant Science > Plant biochemistry > Protein > Modification
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








