- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Transcription (IVT) and tRNA Binding Assay
Published: Vol 4, Iss 18, Sep 20, 2014 DOI: 10.21769/BioProtoc.1234 Views: 16116
Reviewed by: Kanika GeraFanglian He

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Transcriptional Run-on: Measuring Nascent Transcription at Specific Genomic Sites in Yeast
Victoria Begley [...] Sebastián Chávez
Jun 20, 2021 3139 Views

An in vitro Assay of mRNA 3’ end Using the E. coli Cell-free Expression System
Monford Paul Abishek N and Heon M. Lim
Feb 20, 2022 3681 Views

TGIRT-seq Protocol for the Comprehensive Profiling of Coding and Non-coding RNA Biotypes in Cellular, Extracellular Vesicle, and Plasma RNAs
Hengyi Xu [...] Alan M. Lambowitz
Dec 5, 2021 7288 Views
Abstract
This protocol describes the coupling of (i) “live” in vitro RNA transcription with (ii) binding by a radiolabeled, pre-formed tRNA followed by native gel electrophoresis and phosphorimager scan to visualize the complex. The necessity arose from the stable structure that one RNA forms in the absence of its interaction partner. The T-box leader RNA, a transcription control system, folds into a thermodynamically very stable stem-loop structure without the tRNA present, which makes in vitro binding interaction of both pre-formed RNAs very difficult. I therefore adjusted the binding assay to mimic the “natural” situation in the bacterial cell, where the pre-formed, stable tRNA is already present while the T-box leader RNA is actively transcribed by the RNA polymerase. The first part of the protocol also describes the in vitro transcription and labeling of the tRNA.
Keywords: T-boxMaterials and Reagents
- For in vitro transcription (IVT) in general
- T7 RNA polymerase including 5x T7 transcription buffer (Thermo Fisher Scientific, Fermentas, catalog number: EP0111 )
- RiboLock RNase inhibitor (Thermo Fisher Scientific, Fermentas, catalog number: EO0381 )
- 100 mM of ATP, UTP, GTP, CTP (NTP set) (Thermo Fisher Scientific, Fermentas, catalog number: R0481 )
- Recombinant DNase I (Life Technologies, Ambion®, catalog number: AM2235 )
- RNase-free water (Life Technologies, Ambion®, catalog number: AM9932 )
- [alpha 32P]-CTP (800 Ci/mmol, 10 µCi/µl) (PerkinElmer, catalog number: BLU008X )
- 1x T7 transcription buffer (see Recipes)
Note: Templates will be generated by PCR, so all reagents for PCR are needed, too.
- T7 RNA polymerase including 5x T7 transcription buffer (Thermo Fisher Scientific, Fermentas, catalog number: EP0111 )
- For tRNA IVT in particular
- MinElute PCR Purification Kit (QIAGEN, catalog number: 28004 )
- Guanosine 5’-monophosphate disodium salt hydrate from yeast (GMP) (as 100 mM stock solution in RNase-free water) (Sigma-Aldrich, catalog number: G8377 )
- Bio-Spin chromatography columns, Bio-Gel P-6 in Tris buffer (Bio-Rad Laboratories, catalog number: 732-6227 )
- OptiPhase Supermix liquid scintillation cocktail (PerkinElmer, catalog number: 1200-439 )
- Microbeta Starter kit plates (PerkinElmer, catalog number: 1450-486 )
- MinElute PCR Purification Kit (QIAGEN, catalog number: 28004 )
- For denaturing Urea-PAGE
- Urea (Sigma-Aldrich, catalog number: U6504 )
- Rotiphorese 40% acrylamide/bis-acrylamide (19:1) solution (Carl Roth, catalog number: 3030.1 )
- Ammonium persulfate (APS) (Sigma-Aldrich, catalog number: 215589 ) [as 10% (w/v) solution in RNase-free water]
- N,N,N’,N’-Tetramethylethylenediamine (TEMED) (Sigma-Aldrich, catalog number: T9281 )
- 2x MOPS buffer (e.g. Life Technologies, Ambion®, catalog number: AM9570 )
- Formamide (e.g. Life Technologies, Ambion®, catalog number: AM9342 )
- Formaldehyde (e.g. Sigma-Aldrich, catalog number: F8775 )
- Saccharose (e.g. Sigma-Aldrich, catalog number: S7903 )
- Bromophenol blue (e.g. Sigma-Aldrich, catalog number: B0126 )
- Xylencyanol (e.g. Sigma-Aldrich, catalog number: X4126 )
- 1x TBE buffer (see Recipes)
- 2x RNA loading dye (see Recipes)
- Urea (Sigma-Aldrich, catalog number: U6504 )
- For native polyacrylamide gel electrophoresis (PAGE)
- 40% acrylamide/bis-acrylamide (19:1) solution, APS and TEMED as listed in C
- Glycerol (e.g. Sigma-Aldrich, catalog number: G5516 )
- Tris (e.g. Sigma-Aldrich, catalog number: T1503 )
- Boric acid (e.g. Sigma-Aldrich, catalog number: B7901 )
- EDTA (e.g. Sigma-Aldrich, catalog number: E9884 )
- 0.5x TBE buffer (see Recipes)
- 10x loading buffer (see Recipes)
- 40% acrylamide/bis-acrylamide (19:1) solution, APS and TEMED as listed in C
Equipment
- Basic lab equipment for molecular biology with emphasis on RNase-free
- 0.5 ml RNase-free microfuge tubes (Applied Biosystems®, catalog number: AM12300 )
- 1.5 ml RNase-free microfuge tubes (Applied Biosystems®, catalog number: AM12400 )
- Microliter pipettes (e.g. Eppendorf) with tips and 15 ml or 50 ml tubes (e.g. SARSTEDT AG)
- Table-top centrifuge (e.g. Microcentrifuge, Eppendorf, catalog number: 5425 )
- Heating block for 1.5 ml and 0.5 ml tubes (e.g. Grant Instruments, Grant bio PCH-1 personal benchtop cooler/heaters)
Note: Templates will be generated by PCR, so a standard PCR thermo cycler is needed, too.
- 0.5 ml RNase-free microfuge tubes (Applied Biosystems®, catalog number: AM12300 )
- Lab equipment in designated area for radioactive work
- Liquid scintillation counter (PerkinElmer, model: 1450 LSC & Luminescence counter)
- Electrophoresis chamber, glass slides, spacers, clamps and combs for vertical PAGE (e.g. C.B.S. Scientific, catalog number: WSP2-SG-200 ), power supply (e.g. C.B.S. Scientific, catalog number: EPS-200-X)
- Plastic wrap, transparent sheets and Whatman paper (e.g. Sigma-Aldrich, catalog number: Z742422 )
- Gel dryer (Model 583 Gel Dryer) (Bio-Rad Laboratories, catalog number: 165-1746 ) with vacuum pump
- PhosphorImager (Fujifilm FLA-7000) (GE Healthcare, catalog number: 28-9558-09 ) and PhosphorImager screen (storage phosphor screens) (VWR International, catalog number: 28-9564 ) in cassette (VWR International, catalog number: 63-0035 )
- Liquid scintillation counter (PerkinElmer, model: 1450 LSC & Luminescence counter)
Procedure
- In vitro transcription (IVT) of tRNAs with free 3’cca end
- Primer for amplification of the tRNA template from the bacterial chromosome by PCR need the T7 promoter sequence at their 5’ end for in vitro transcription by the T7 polymerase. For successful transcription, three purine bases, at best guanine, should follow the core promoter sequence (underlined): nnnnCTAATACGACTCACTATAGRRnnnnnn.… with the first G (in bold) as transcription start.
- PCR products for tRNA templates are cleaned up with the MinElute PCR Purification Kit according to the manufacturer’s protocol.
- The tRNA IVT reaction is set up in 20 µl volume as follows.
- Add 1x T7 reaction buffer, 20 U RiboLock™ RNase inhibitor, 0.5 mM ATP, GTP and UTP, 12 µM CTP, 9 mM GMP, 20 U T7 RNA polymerase and 2 µl of [alpha 32P]-CTP (800 Ci/mmol, 10 µCi/µl) to 4 pmol of DNA template (tRNA-PCR).
Note: The reaction set up follows the protocol from Fermentas for Synthesis of Radiolabeled RNA Probes of High Specific Activity (Fermentas). The GMP is added to increase the 5’ monophosphorylation of the resulting tRNA molecules (according to Sampson and Uhlenbeck, 1988). - Incubation of the reaction mix at 37 °C for <7 h to enrich transcription of short fragments and stop of reaction at -20 °C for 5 min.
- DNA template is removed by adding 1 U of recombinant DNase I and incubating at 37 °C for further 30 min.
- tRNA products have to be cleaned up using e.g. the Micro Bio-Spin™ chromatography columns with Bio-Gel P-6 in Tris buffer to ensure no small tRNA fragments are lost. Keep 1 µl of the reaction volume before the clean-up if you use the scintillation counter to measure the incorporation rate (see A4).
Note: Other clean up kits are of course possible as long as loss of the desired product is avoided and unincorporated, radioactive nucleotides are removed. If more than one tRNA product can be detected by denaturing Urea-PAGE (see Figure 1), the correct band should rather be eluted from the gel. The recipe for denaturing Urea-PAGE can be found after this section.
- Add 1x T7 reaction buffer, 20 U RiboLock™ RNase inhibitor, 0.5 mM ATP, GTP and UTP, 12 µM CTP, 9 mM GMP, 20 U T7 RNA polymerase and 2 µl of [alpha 32P]-CTP (800 Ci/mmol, 10 µCi/µl) to 4 pmol of DNA template (tRNA-PCR).
- To establish the incorporation rate of radionucleotides during IVT into the produced tRNA, 1 µl of the reaction volume before and after clean-up is used for scintillation count.
- OptiPhase Supermix liquid scintillation cocktail (50 µl per well) are pipetted into a rigid 96-well plate and the 1 µl samples are added, with one empty well between samples to avoid cross-counting of wells.
- In parallel, a dilution series of the utilized radionucleotide [alpha 32P]-CTP is added to the same plate to obtain a standard curve for the cpm (counts per min).
- The plate is sealed and placed in the liquid scintillation counter. The cpm from the standard curve will give the cpm value per µCi. This will change with each experiment, due to the half life of P32. The cpm values of the sample before and after clean-up helps to calculate the incorporation rate of radionucleotides and subsequently the RNA yield after IVT to determine the molarity of tRNA to be used in the binding assay.
Note: Incorporation rate and RNA yield can be calculated with help of an article by Ambion (http://www.lifetechnologies.com/de/de/home/references/ambion-tech-support/nuclease-enzymes/tech-notes/determining-rna-probe-specific-activity-and-yield.html). If no scintillation counter is available, incorporation rate can also be estimated through Geiger counter values obtained before and after clean-up and the RNA yield and molarity calculated from there.
- OptiPhase Supermix liquid scintillation cocktail (50 µl per well) are pipetted into a rigid 96-well plate and the 1 µl samples are added, with one empty well between samples to avoid cross-counting of wells.
- Primer for amplification of the tRNA template from the bacterial chromosome by PCR need the T7 promoter sequence at their 5’ end for in vitro transcription by the T7 polymerase. For successful transcription, three purine bases, at best guanine, should follow the core promoter sequence (underlined): nnnnCTAATACGACTCACTATAGRRnnnnnn.… with the first G (in bold) as transcription start.
- Recipe for denaturing Urea-PAGE
- A 10% (w/v) acrylamide/ 8 M urea gel was chosen for the separation of 78 nt tRNA molecules on a small size gel (10 x 10 cm).
- 3.36 g urea are solved in 1x TBE (up to volume of 5.25 ml) by stirring for > 4 h at room temperature. 1.75 ml 40% acrylamide/bis-acrylamide (19: 1) solution are added to the 8 M urea solution to reach 7 ml total volume.
Note: The total volume depends on size of glass plates and thickness of spacers used. Here we work with 10 x 10 cm plates and 1 mm thickness and need 7 ml volume. The urea-acrylamide mix can be prepared in advance in a bigger volume and stored at 4 °C. - Clean glass slides of desired size are set up with spacer on left and right and either put in a gel casting chamber or fixed with clamps on both sides.
- 35 μl [0.5% (v/v)] of 10% APS and 15 μl 0.2% (v/v) TEMED are added to the urea-acrylamide mix to catalyze the polymerization process.
- The mix is quickly poured between the glass slides (e.g. with 10 ml glass pipette) and the comb with desired amount of wells adjusted in the gel. Avoid air bubbles!
- 3.36 g urea are solved in 1x TBE (up to volume of 5.25 ml) by stirring for > 4 h at room temperature. 1.75 ml 40% acrylamide/bis-acrylamide (19: 1) solution are added to the 8 M urea solution to reach 7 ml total volume.
- The fully polymerized gel was pre-run in 1x TBE buffer at 250 V at room temperature for 40 min. Remove the comb for that.
- tRNA samples were mixed with 2x RNA loading dye and heat denatured at 85 °C for 5 min before loading.
Note: Rinse the wells thoroughly right before loading the samples! - Electrophoresis settings are 250 V at room temperature to run for 1 h in 1x TBE buffer.
- After electrophoresis run the gel is transferred to Whatman paper and covered in a transparent sheet before exposure of a PhosphorImager screen (couple of hours is usually sufficient), followed by scan with the PhosphorImager FLA-7000 (see Figure 1).
Note: Do not dry Urea-PA gels with the gel dryer, as they will disintegrate.
- A 10% (w/v) acrylamide/ 8 M urea gel was chosen for the separation of 78 nt tRNA molecules on a small size gel (10 x 10 cm).
- IVT-RNA-tRNA binding assay
- A typical 10 μl reaction volume contains all components for T7 in vitro transcription:
- 1x T7 transcription buffer, 10 U of RiboLock RNase inhibitor, 6 U of T7 RNA polymerase and 0.5 mM NTPs (ATP, UTP, GTP, CTP). DNA template (PCR product) for the leader RNA is added to 8 nM and the radiolabeled tRNA to 50 nM.
Note: The leader RNA template has to contain a T7 promoter sequence at the 5’ end as described in A1. - Control reactions are set up accordingly without tRNA but with 12 µM CTP and 1 µl [alpha 32P]-CTP (800 Ci/mmol, 10 µCi /µl) to check for IVT efficiency.
- The reaction mix is incubated at 37 °C for 2 h, which is sufficient for the 440 nt transcript.
- 2 µl of 10x loading buffer are added and the samples immediately loaded onto a non-denaturing polyacrylamide gel.
- 1x T7 transcription buffer, 10 U of RiboLock RNase inhibitor, 6 U of T7 RNA polymerase and 0.5 mM NTPs (ATP, UTP, GTP, CTP). DNA template (PCR product) for the leader RNA is added to 8 nM and the radiolabeled tRNA to 50 nM.
- A 6% (w/v) acrylamide gel was chosen to separate a 78 nt tRNA from the complex with a ~440 nt RNA on a medium size vertical gel.
- Clean glass slides of desired size are set up with spacer on left and right and either put in a gel casting chamber or fixed with clamps on both sides.
- 1.5 ml 40% acrylamide/bis-acrylamide (19:1) solution and 0.5 ml 10x TBE are mixed with RNase-free water to 10 ml volume and then 40 μl [0.4% (v/v)] of 10% APS and 20 μl [0.2% (v/v)] TEMED are added to catalyze the polymerization process.
Note: The total volume depends on size of glass plates and thickness of spacers used. Here we work with 10 x 15 cm plates and 1 mm thickness and need 10 ml volume. Higher percentage of TEMED than usual was used to achieve a very quick polymerization. - The mix is quickly poured between the glass slides (e.g. with 10 ml glass pipette) and the comb with desired amount of wells adjusted in the gel. Avoid air bubbles!
- The fully polymerized gel has to be pre-run in 0.5x TBE buffer at 200 V at 4 °C for at least 40 min. Remove the comb for that. Rinse the wells before loading samples.
- Immediately load the samples after binding reaction.
Note: If possible, load the samples quickly while electrophoresis chamber is already running to avoid disassociation of the complex. - Electrophoresis settings are 100 V (= 6.7 V/cm), 5 W at 4 °C to run for >3 h.
Note: These settings depend mainly on the size difference between unbound and bound labeled RNA moiety and on the size of the gel used (so it should not run out the bottom but be clearly separated). It is much better to cool the gel while running (so either in a 4 °C room or with a buffer cooling system).
- Clean glass slides of desired size are set up with spacer on left and right and either put in a gel casting chamber or fixed with clamps on both sides.
- After electrophoresis, the gel is transferred onto a Whatman paper and covered with plastic wrap, then dried by vacuum gel dryer at 80 °C for < 30 min.
- Detection of the radiolabeled RNA complex is achieved by exposure of a PhosphorImager screen (often over night), followed by scan with the PhosphorImager FLA-7000 (see Figure 2).
- A typical 10 μl reaction volume contains all components for T7 in vitro transcription:
Representative data
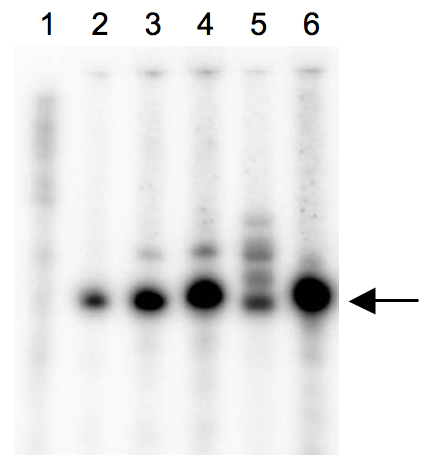
Figure 1. Scanned image of denaturing Urea-PAGE of labelled tRNAs. This example shows dominant single products for most tRNA IVT reactions (arrow), but also a diffuse pattern of several products in one sample (lane 5). This sample would result in an unspecific binding reaction, thus the IVT must be repeated or the correct band eluted from the gel. lane 1: size marker; lane 2 - 6: different in vitro transcribed tRNAs.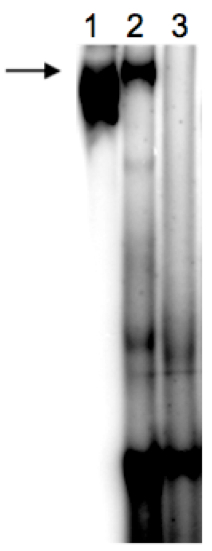
Figure 2. Scanned image of native PAGE gel after binding assay. This is an example of the separation of unbound, small tRNA* at the bottom of the gel and the tRNA* bound to the much bigger T-box RNA (arrow), which was transcribed in vitro in the presence of pre-formed, radiolabelled(*) tRNA. lane 1: control IVT reaction of T-box RNA* without tRNA; lane 2: T-box RNA IVT with tRNA*fMet; lane 3: T-box RNA IVT with tRNA*Cys (not binding to T-box RNA).
Notes
- The most variable component in our hands was the in vitro transcription of the tRNA molecules. Only if one dominant tRNA product was achieved [controlled by denaturing PAGE with 10 % (w/v) acrylamide/ 8 M urea, see Figure 1], then the detected binding interaction was strong. If one discovers a high diversity in the produced tRNA moiety, one might be better advised to elute the labelled tRNA band from the urea-acrylamide gel before the binding assay.
Recipes
- 5x T7 transcription buffer (usually supplied with T7 polymerase)
200 mM Tris/HCl (pH 7.9)
50 mM DTT
30 mM MgCl2
50 mM NaCl
10 mM spermidine - 10x TBE buffer (1 L)
0.89 M Tris
0.9 M boric acid
25 mM EDTA
Solve 108 g Tris and 55 g boric acid in 950 ml distilled water
Add 50 ml 0.5 M EDTA (pH 8.0) - 2x RNA loading dye
2x MOPS buffer
65% (v/v) formamide
4.4% (v/v) formaldehyde
2% (w/v) saccharose
0.1% (w/v) bromophenol blue
0.1% (w/v) xylencyanol - 10x loading buffer
40% (v/v) glycerol
0.25x TBE
Acknowledgments
This work was funded by the German Research Foundation (DFG), Transregio34 “Pathophysiology of staphylococci in the post-genomic era”, and my PhD stipend was granted by the European Social Fund (ESF). The protocol presented here was developed in the lab of PD Dr. Wilma Ziebuhr during my research time at the Queen’s University Belfast (QUB).
References
- Fermentas T7 IVT protocols: http://www.thermoscientificbio.com/dna-and-rna-modifying-enzymes/t7-rna-polymerase/
- Sampson, J. R. and Uhlenbeck, O. C. (1988). Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A 85(4): 1033-1037.
- Schoenfelder, S. M., Marincola, G., Geiger, T., Goerke, C., Wolz, C. and Ziebuhr, W. (2013). Methionine biosynthesis in Staphylococcus aureus is tightly controlled by a hierarchical network involving an initiator tRNA-specific T-box riboswitch. PLoS Pathog 9(9): e1003606.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Schoenfelder, S. M. (2014). In vitro Transcription (IVT) and tRNA Binding Assay. Bio-protocol 4(18): e1234. DOI: 10.21769/BioProtoc.1234.
- Schoenfelder, S. M., Marincola, G., Geiger, T., Goerke, C., Wolz, C. and Ziebuhr, W. (2013). Methionine biosynthesis in Staphylococcus aureus is tightly controlled by a hierarchical network involving an initiator tRNA-specific T-box riboswitch. PLoS Pathog 9(9): e1003606.
Category
Microbiology > Microbial genetics > RNA > Transcription
Molecular Biology > RNA > Transcription
Molecular Biology > RNA > RNA labeling
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









