- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A SYBR Green-based Real Time RT-PCR Assay for Detection of the Emerging H7N9 Virus
Published: Vol 4, Iss 12, Jun 20, 2014 DOI: 10.21769/BioProtoc.1150 Views: 10098
Reviewed by: Vinay PanwarLee-Hwa TaiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Infectious Virus Yield Assay for Hepatitis E Virus
Yannick Debing [...] Johan Neyts
Aug 5, 2014 11471 Views

Quantification of HIV RNA and Human Herpesvirus DNA in Seminal Plasma
Milenka V. Vargas-Meneses [...] Sara Gianella
May 5, 2015 11124 Views

Detection of Cytoplasmic and Nuclear Circular RNA via RT-qPCR
Ke-En Tan [...] Yat-Yuen Lim
Sep 5, 2023 3392 Views
Abstract
Most recently a novel avian-origin influenza A (H7N9) virus emerged in China and has been associated with lots of human infection and fatal cases. Molecular diagnostic methods are thus urgently needed in public health laboratories. We developed a SYBR green-based one-step real time reverse transcription-PCR (RT-PCR) to detect the novel H7N9 virus.
Materials and Reagents
- Respiratory specimens (throat-swabs, sputum and tracheal aspirate) from patients with influenza-like illnesses
- Madin-Darby canine kidney (MDCK) cells (Shanghai Institutes for Biological Sciences)
- Eagle's Minimum Essential Medium (EMEM) (Life Technologies, Gibco®, catalog number: 11095-080 )
- Fetal bovine serum (Life Technologies, Gibco®, catalog number: 10099 )
- Viral transportation medium (Yocon Bio-technology, catalog number: MT0301-1 )
- QIAsymphony Virus/Bacteria Mini Kit (QIAGEN, catalog number: 931036 )
- Nuclease-free water (Life Technologies, catalog number: 10977 )
- SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Life Technologies, catalog number: 11736 )
- The primer sequences used for detection of influenza A H7N9 virus
a F: Forward; R: ReversePrimera Sequence (5’-3’)b PCR amplicon (bp) H7FH7RN9FN9RTGAAAATGGVTGGGAAGGYYTGCCGATTGRGTGCTYTTRTACAGTGTACAAYAGCARRGTGTTTCGRGCCCAYGTRTTAA103165
b V: A/C/G; Y: C/T; R: A/G
Equipment
- T-25 cell culture flask (Corning, catalog number: 430168 )
- A biosafety 3 laboratory (BSL-3) (required for virus isolation)
- QIAsymphony SP instrument (QIAGEN, catalog number: 9001297 )
- ABI Prism 7900HT Sequence Detection System (Life Technologies, Applied Biosystems®, catalog number: 4329001 )
Software
- Sequence Detection System Software (v2.3) (Life Technologies, Applied Biosystems®)
Procedure
- Virus isolation
- MDCK cells were cultured in Eagle's Minimum Essential Medium (EMEM) containing 10% heat-inactivated fetal bovine serum (FBS) in a T-25 cell culture flask and incubated at 37 °C under a humidified atmosphere with 5% CO2.
- Disperse the respiratory specimens (throat-swabs, sputum and tracheal aspirate) maintained in the viral transportation medium by vortex for 1 min. Sputum was pre-diluted with an equal volume of PBS solution prior to a vortex procedure.
- When the MDCK cells grew to 85% confluence, 0.5 ml of the respiratory specimens positive for H7N9 virus infection was inoculated into cells.
- Cytopathic effects were observed every day by microscopy.
- When 90% of MDCK cells experienced cytopathic effects (Figure 1), the cell culture supernatants were harvested by centrifuge at 2,000 rpm for 10 min and stored at -70 °C until use.
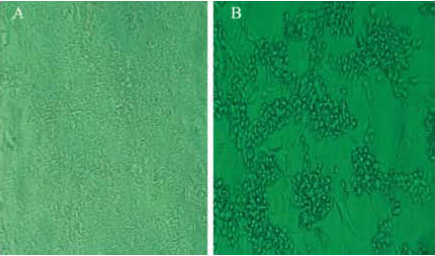
Figure 1. Rounding of infected cells, fusion with adjacent cells to form a syncytia and ultimate lysis, as compared with non-infected cells. A: Non-infected cells; B: Cells with cytopathic effects. - 200 μl of cell culture supernatants was subjected to RNA extraction. Purified viral nucleic acid was used as assay control.
- MDCK cells were cultured in Eagle's Minimum Essential Medium (EMEM) containing 10% heat-inactivated fetal bovine serum (FBS) in a T-25 cell culture flask and incubated at 37 °C under a humidified atmosphere with 5% CO2.
- RNA extraction
- Viral RNA was extracted from 200 μl of cell culture supernatants or respiratory specimens using QIAsymphony Virus/Bacteria Mini Kit combined with the QIAsymphony SP instrument, following the manufacturer’s instructions.
- The extracted RNA was eluted in 60 μl of nuclease-free water. RNA integrity was analyzed by agarose gel electrophoresis and then stored at -70 °C until use.
- Viral RNA was extracted from 200 μl of cell culture supernatants or respiratory specimens using QIAsymphony Virus/Bacteria Mini Kit combined with the QIAsymphony SP instrument, following the manufacturer’s instructions.
- SYBR green-based real time RT-PCR assay
- The one-step real time quantitative RT-PCR assays were performed to amplify the HA and NA genes of H7N9 virus respectively using SuperScript III Platinum SYBR Green One-Step qRT-PCR kit. The assay was run in duplicate or triplicate for each unknown sample.
- The assay was carried out in a 10 μl reaction mixture containing:
5 μl of 2x SYBR Green Reaction Mix
0.8 μM of each primer
0.2 μl of ROX Reference Dye (500 nM)
0.2 μl of SuperScript III RT/Platinum Taq Mix
1 μl of purified RNA
x μl of nuclease-free water - The optimized thermal cycling conditions were as follows:
- A reverse transcription step at 50 °C for 10 min
- An initial denaturation step at 95 °C for 5 min
- 40 cycles of PCR amplification at 95 °C for 15 sec, 60 °C for 20 sec, and 72 °C for 30 sec, followed by a melting curve analysis program according to the instrument documentation.
- A reverse transcription step at 50 °C for 10 min
- Finally, data were collected and results were analyzed with the use of Sequence Detection System Software v2.3. The Tm values of H7 and N9 specific amplicons were 80.77 and 81.20, respectively.
- The one-step real time quantitative RT-PCR assays were performed to amplify the HA and NA genes of H7N9 virus respectively using SuperScript III Platinum SYBR Green One-Step qRT-PCR kit. The assay was run in duplicate or triplicate for each unknown sample.
Acknowledgments
This experimental protocol was partly adapted from the previously published paper: Zhu et al. (2013). This protocol was supported in part by the Jiangsu Province Health Development Project with Science and Education (ZX201109), the Jiangsu Province Key Medical Talent Foundation (JKRC2011002, RC2011191), and the Science and Technology Pillar program of Jiangsu Province (BE2011796). We thank Dr. Shu Yuelong, China National Influenza Center, Chinese Center for Disease Control and Prevention, Beijing, China, for providing the entire gene sequence data of the first three H7N9 virus isolates used for primer design.
References
- Zhu, Z., Fan, H., Qi, X., Qi, Y., Shi, Z., Wang, H., Cui, L. and Zhou, M. (2013). Development and evaluation of a SYBR Green-based Real Time RT-PCR assay for detection of the emerging avian influenza A (H7N9) birus. PLoS One 8(11): e80028.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhu, Z. and Cui, L. (2014). A SYBR Green-based Real Time RT-PCR Assay for Detection of the Emerging H7N9 Virus. Bio-protocol 4(12): e1150. DOI: 10.21769/BioProtoc.1150.
Category
Microbiology > Microbial genetics > RNA > qRT-PCR
Molecular Biology > RNA > qRT-PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









