- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Library Construction for Genome-wide Bisulfite Sequencing in Plants
Published: Vol 3, Iss 24, Dec 20, 2013 DOI: 10.21769/BioProtoc.1013 Views: 15130
Reviewed by: Feng Li

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Chromosome Dosage Analysis in Plants Using Whole Genome Sequencing
Ek Han Tan [...] Isabelle M. Henry
Jul 5, 2016 12934 Views

DNA-free Genome Editing of Chlamydomonas reinhardtii Using CRISPR and Subsequent Mutant Analysis
Jihyeon Yu [...] Sangsu Bae
Jun 5, 2017 16000 Views

Tethered Chromosome Conformation Capture Sequencing in Triticeae: A Valuable Tool for Genome Assembly
Axel Himmelbach [...] Nils Stein
Aug 5, 2018 9900 Views
Abstract
DNA methylation is the most studied epigenetic modification, which involves the addition of a methyl group to the carbon-5 position of cytosine residues in DNA. DNA methylation is important for the regulation of gene expression. Bisulfite sequencing is the gold standard technique for determining genome-wide DNA methylation profiles in eukaryotes. This protocol describes how to prepare libraries of genomic DNA for whole-genome bisulfite sequencing in Arabidopsis, which could be adapted for use in other plant species.
Materials and Reagents
- DNA LoBind 1.5 and 2.0 ml centrifuge tubes (Eppendorf, catalog number: 22431021 and 22431048 , respectively)
- AMPure magnetic beads (Beckman Coulter, catalog number: A63881 )
- Ethanol (70% v/v, ethanol/water)
- TruSeq DNA sample preparation kit (Illumina, catalog number: FC-121-2001 )
- SYBR Safe (Life Technologies, catalog number: S33102 )
- 50 bp ladder (Thermo Fisher Scientific, catalog number: SM0373 )
- Disposable sterile scalpels (Thermo Fisher Scientific catalog number 12-460--452 )
- FalconTM 15 ml conical centrifuge tubes (Thermo Fisher Scientific, catalog number: 14-959-49B )
- QIAquick gel extraction kit (QIAGEN, catalog number: 28704 )
- Qubit dsDNA HS assay kit (Life Technologies, catalog number: Q32851 )
- EpiTect bisulfite kit (QIAGEN, catalog number: 59104 )
- MinElute PCR purification kit (QIAGEN, catalog number: 28004 )
- PfuTurbo Cx hotstart DNA polymerase (Agilent, catalog number: 600410-51 )
- KAPA library quantification kit (Kapa Biosystems, catalog number: KK4824 )
- TE buffer (see Recipes)
- Elution buffer (see Recipes)
Equipment
- S2 adaptive focus acoustic disruptor (Covaris)
- Microtubes with AFA fiber with pre-slit snap-caps (Covaris, catalog number: 520045 )
- Refrigerated water bath circulator (Thermo Fisher Scientific, model: SC100-A10 )
- Thermomixer R (Eppendorf, catalog number: 022670107 )
- MiniSpin PlusTM centrifuge (Eppendorf, catalog number 0 22620100 )
- Magnetic stand (Life Technologies, catalog number 4457858 )
- Vortex mixer (Thermo Fisher Scientific, catalog number 02-215-365 )
- 2100 BioAnalyzer (Agilent, catalog number G2943CA )
- Qubit 2.0 fluorometer (Life Technologies, catalog number Q32866 )
- Horizon 11-14, horizontal gel electrophoresis system (Apogee Designs, model: 11068020 )
- Safe Imager 2.0 blue light transilluminator (Life Technologies, model: G6600 )
- ABI7900HT real-time PCR system (Life Technologies)
- Thermal cycler (C1000 Touch) (Bio-Rad Laboratories, model: 185-1196EDU )
- Pipettors (RAININ, various models)
Procedure
The overall workflow of the procedure is illustrated in Figure 1.
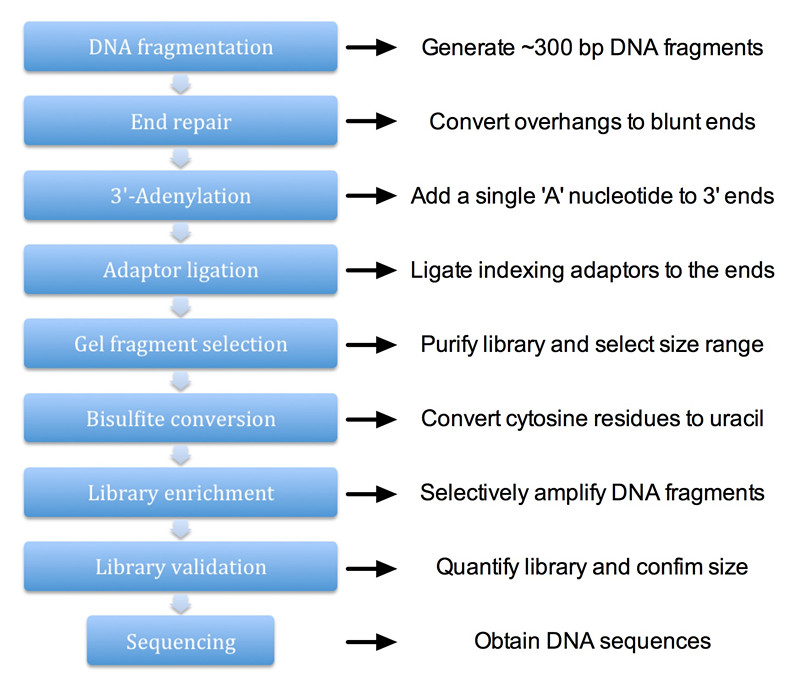
Figure 1. Workflow of library construction for genome-wide bisulfite sequencing
- DNA fragmentation by ultrasonic destruction
- Dissolve ~2 μg of high quality, high molecular weight genomic DNA extracted from Arabidopsis leaf tissues in 130 μl TE buffer and transfer to a 6 x 16 mm glass microtube with AFA fiber with pre-slit snap-caps. Genomic DNA extracted using the commonly used CTAB (cetyltrimethyl ammonium bromide) method is suitable as long as the genomic DNA runs at a high molecular weight (> 12 kb) and as a clean single band in 0.8% agarose gel. The quality of genomic DNA can also be monitored with the 2100 BioAnalyzer (Figure 2A).
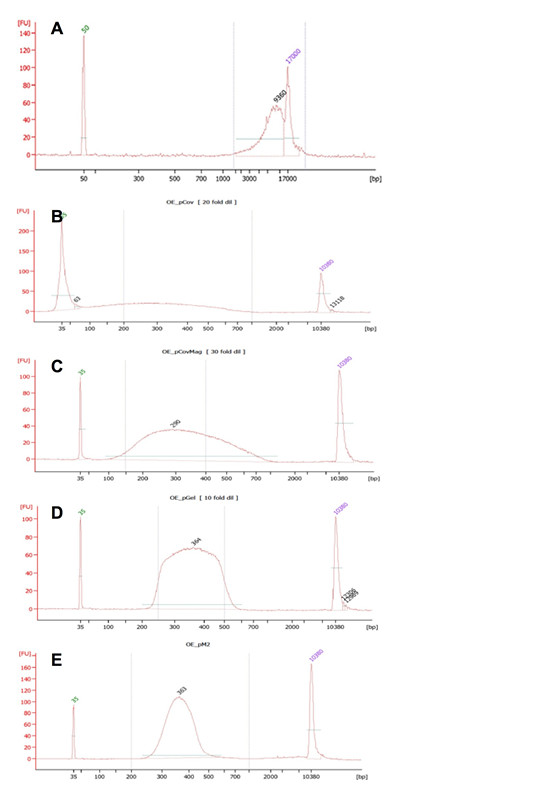
Figure 2. BioAnalyzer traces of an Arabidopsis DNA sample (OE) that was processed through the workflow of the procedure described in this protocol. A. Genomic DNA; B. Ultrasonically fragmented DNA; C. Ultrasonically fragmented and magnetic bead cleaned up DNA; D. Adaptor-ligated DNA fragments recovered by the agarose gel-based selection; E. The final sequencing library (post PCR amplification).
- Shear the genomic DNA into fragments with an average size of 300 bp in the S2 adaptive focus acoustic disruptor using the following settings (for fragmenting DNA with the majority of fragments in the 150-450 bp range) (Figure 2B):
Duty cycle 10%
Intensity 4
Cycles/burst 200
Time 132 sec
Temperature 6 °C
The S2 instrument is connected to the refrigerated water bath circulator that is set at 6 °C in order to maintain a constant temperature during the sonication process.
- Transfer to a 1.5 ml microcentrifuge tube, and estimate the recovered volume with a pipettor. Typically, 125 μl of the DNA solution is recovered.
- Add a suspension (slur) of AMPure magnetic beads at a bead-to-sample ratio of 0.73:1 (v/v). This bead-to-sample ratio is used to collect DNA fragments over 200 bp prior to construction of the sequencing library.
- Incubate the mix at room temperature for 15 min with shaking at 700 rpm in the Thermomixer R to allow DNA to bind with the beads.
- Pulse spin to collect the mix at the bottom of the tube, and then put the tube on the magnetic stand for 1-2 min until the solution is completely clear. The magnetic beads will be attracted to the magnet, thus allowing removal of the supernatant. Remove and discard the supernatant.
- Leave the tube on the magnetic stand and wash the beads twice as follows: for each wash add 1 ml of freshly prepared 70% ethanol, gently pipet the beads up and down six times, let the beads sit for 30-60 sec, and then discard the supernatant by pipetting without disturbing the beads.
- Following the second wash, pulse spin the tube to collect traces of ethanol, and then place the tube on the magnetic stand. Remove ethanol traces with a fine-tipped pipettor.
- Air dry the magnetic beads for 1 to 3 min at room temperature.
- Elute DNA as follows: remove the tube from the magnetic stand and place on a plastic rack, add 30 μl of elution buffer, vortex vigorously for 10 sec, place the tube back on the magnetic stand until the solution becomes completely clear, and then use a pipettor to transfer the supernatant containing the DNA into a clean microcentrifuge tube. Repeat the elution by adding another 30 μl of elution buffer as above. Pool the two eluate fractions.
- Save 3 μl of the DNA sample for quantification in the Qubit 2.0 fluorometer according to the product instructions (http://www.lifetechnologies.com/order/catalog/product/Q32854) and for fragment size confirmation with the 2100 BioAnalyzer (Figure 2C). The desired size range of the fragments should be 200-450 bp.
At this point the DNA can be stored at -20 °C for later use.
- Dissolve ~2 μg of high quality, high molecular weight genomic DNA extracted from Arabidopsis leaf tissues in 130 μl TE buffer and transfer to a 6 x 16 mm glass microtube with AFA fiber with pre-slit snap-caps. Genomic DNA extracted using the commonly used CTAB (cetyltrimethyl ammonium bromide) method is suitable as long as the genomic DNA runs at a high molecular weight (> 12 kb) and as a clean single band in 0.8% agarose gel. The quality of genomic DNA can also be monitored with the 2100 BioAnalyzer (Figure 2A).
- Library construction
Sequencing libraries are made using the low-throughput TruSeq DNA sample preparation kit following the manufacturer's protocol (TruSeq DNA sample preparation guide) with a few modifications (see below). The adaptors included in this kit are methylated and are therefore compatible with bisulfite sequencing for DNA methylation analysis.
- End repair the DNA fragments in the following reaction:
50 μl above DNA sample (~1.5 μg)
10 μl end-repair reaction buffer (included in the TruSeq Kit)
40 μl end-repair mix (included in the TruSeq Kit)
Incubate at 30 °C for 30 min. Clean up the reaction mix with AMPure magnetic beads as described above (DNA Fragmentation section: steps A4-10), except that the bead-to-sample ratio is 1:1 (v/v). Elute DNA in 17.5 μl elution buffer. This bead-to-sample ratio is used to collect essentially all DNA fragments.
- 3’-adenylate the end-repaired DNA fragments in the following reaction:
15 μl end-repaired DNA
2.5 μl 3'-adenylation reaction buffer (included in the TruSeq Kit)
12.5 μl A-tailing mix (included in the TruSeq Kit)
Incubate at 37 °C for 30 min. The adenylation mix is compatible with the ligation reaction; therefore, no clean-up is necessary at this step.
- Ligate methylated adaptors to the 3'-adenylated DNA fragments in the following reaction:
30 μl adenylated DNA (the entire adenylation reaction from step B2)
2.5 μl ligation reaction buffer (included in the TruSeq Kit)
2.5 μl ligation mix (included in the TruSeq Kit)
2.5 μl adaptors (included in the TruSeq Kit)
Incubate at 30 °C for 10 min. Add 5 μl of ligation stop buffer. Clean-up sample using AMPure magnetic beads as described above (DNA Fragmentation section: steps A4-10). with a 1:1 (v/v) bead-to-sample ratio. Elute adaptor-ligated DNA in 22.5 μl elution buffer.
- Size-select adaptor-ligated DNA fragments (250-500 bp) in a 2% TAE agarose gel as follows. Prepare 100 ml of the solution in a 500 ml Erlenmeyer flask, heat in a microwave oven until agarose is fully dissolved, add SYBR Safe stain (1:10,000 v/v dye-to-gel ratio) to the molten agarose solution and swirl to mix, and then pour the gel-dye solution into properly set-up casting of the gel electrophoresis apparatus. A 10-well comb is placed at one end of the gel casting tray to form wells. Allow gel to fully solidify (~30 min). Load a 50 bp ladder in one of the gel wells to aid in size estimation. Mix the sample with a suitable sample loading dye and load into any of the additional wells. Connect electrodes to power supply and electrophorese at 100 V until the quickest moving dye reaches 3/4 of the gel length from the wells. No post electrophoresis staining is necessary when the dye is added as suggested here. SYBR Safe is used instead of ethidium bromide in order to avoid UV damage (during viewing) of DNA fragments. View the gel on a Safe Imager 2.0 blue light transilluminator. Cut the gel region containing fragments in the 250-500 bp range using a sterile, disposable scalpel (Figure 3). Transfer the gel slice into a DNA LoBind 2.0 ml centrifuge tube. If gel slices are too large, they can be placed in a 15 ml conical tube. After the agarose gel is dissolved (see step 5 below), split dissolved volume if necessary into DNA LoBind 2.0 ml tubes for DNA extraction.
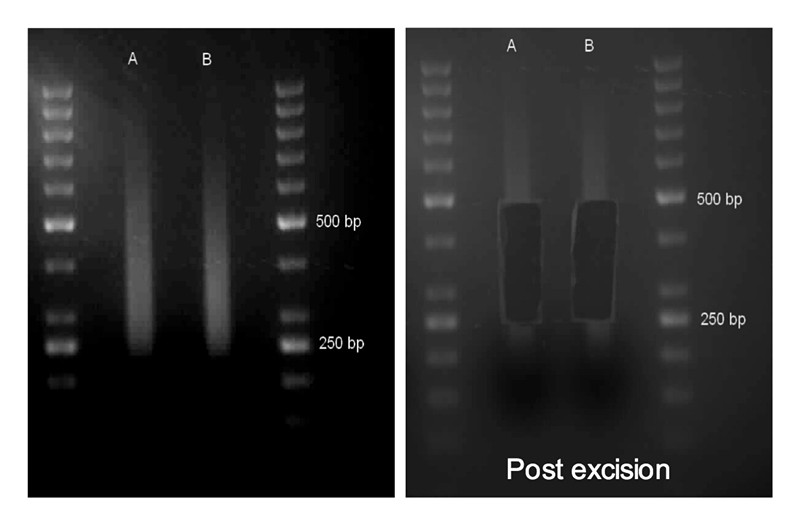
Figure 3. Size selection of adaptor-ligated DNA fragments in an agarose gel. Two Arabidopsis genomic DNA samples (A and B) were fragmented in the S2 adaptive focus acoustic disruptor, end-repaired, 3’-adenylated, adaptor-ligated, and separated in a 2% TAE agarose gel. Gel wells were formed using a 10-well-forming comb (8 mm wide wells). Ten μl of ready-to-load 50 bp ladder were loaded in the flanking positions of the samples in the gel for excision process. Left: before excision; Right: post excision.
- Extract DNA from the gel using the QIAquick gel extraction kit following the manufacturer's instructions (http://www.qiagen.com/products/catalog/sample-technologies/dna-sample-technologies/dna-cleanup/qiaquick-gel-extraction-kit#resources). Elute DNA with 22 μl elution buffer (included in the QIAquick gel extraction kit).
- Set aside 2 μl to quantitate the library in the Qubit 2.0 fluorometer with a Qubit dsDNA HS assay kit according to the product instructions (http://www.lifetechnologies.com/order/catalog/product/Q32854). The final library yield using this protocol is usually 300-400 ng (Figure 2D), which corresponds to a concentration of 13-18 DNA ng/ μl.
At this point the DNA library can be stored at -20 °C for later use.
- End repair the DNA fragments in the following reaction:
- Bisulfite treatment
The entire library (300-400 ng) is subjected to sodium bisulfite treatment using the EpiTect bisulfite kit following the manufacturer's instructions (http://www.qiagen.com/products/catalog/assay-technologies/epigenetics/epitect-bisulfite-kits - resources). Briefly:
- Prepare buffer BW, buffer BD, carrier RNA, buffer BL, and bisulfite mix following the instructions in the kit.
- Treat DNA with bisulfite in the following reaction:
20 μl DNA (the entire library)
85 μl bisulfite mix (included in the EpiTect kit)
35 μl DNA protect buffer (included in the EpiTect kit)
Incubate at the following conditions in a thermal cycler: 95 °C for 5 min, 60 °C for 25 min, 95 °C for 5 min, 60 °C for 85 min, 95 °C for 5 min, 60 °C for 175 min, and finally hold at 20 °C.
- Purify the bisulfite-converted DNA following the manufacturer's instructions. Elute DNA twice, each time with 20 μl elution buffer (included in the EpiTect kit) and pool the two eluate fractions.
- Further purify and concentrate the DNA using a MinElute column following the instructions in the MinElute PCR purification kit (http://www.qiagen.com/products/catalog/sample-technologies/dna-sample-technologies/dna-cleanup/minelute-pcr-purification-kit#resources). Elute DNA in 15 μl elution buffer (included in the MinElute PCR purification kit).
At this point the bisulfite-converted library can be stored at -20 °C for later use.
- Prepare buffer BW, buffer BD, carrier RNA, buffer BL, and bisulfite mix following the instructions in the kit.
- Library enrichment
- Amplify ~1 ng of the bisulfite-converted library using a uracil-insensitive PfuTurbo Cx hotstart DNA polymerase and the primer cocktail included in the Illumina TruSeq library construction kit in the following reaction:
17.25 μl double-distilled water
15 μl DNA template (~1 ng, usually the entire eluate of the above library)
5 μl Pfu Turbo Buffer (coming with the Pfu Turbo polymerase)
2.5 μl DMSO
1.25 μl dNTPs (10 mM)
5 μl primer cocktail (included in the TruSeq Kit)
1μl Pfu Turbo Cx Hotstart polymerase
The amplification conditions are: denaturation at 98 °C for 30 sec, 14-18 cycles (98 °C for 15 sec, 60 °C for 30 sec, and 72 °C for 1 min), and final extension at 72 °C for 5 min. Depending on the yield of the bisulfite treatment step, the number of PCR amplification cycles should be as small as possible. This step is to selectively enrich DNA library fragments that have methylated adaptors at both ends, and to generate sufficient DNA for accurate quantitation.
- Clean the PCR amplification products twice using AMPure magnetic beads as describe above (DNA Fragmentation section: steps A4-10) with a bead-to-sample ratio of 0.85:1 (v/v). Elute DNA in 30 μl of elution buffer. This bead-to-sample ratio is used to remove dNTPs and primers while retaining all the DNA amplification fragments above 150 bp.
- Use 1 μl of the DNA sample for fragment size confirmation with the 2100 BioAnalyzer. The average insert size of the library generated by this protocol is ~360 bp (Figure 2E). A different average size can be obtained by adjusting the DNA ultrasonic fragmentation conditions (DNA fragmentation section: step A2) and by choosing the desired range at the size selection step (Library construction section: step B4).
- Use 1 μl of the DNA sample for quantification in the Qubit 2.0 fluorometer and 1 μl for quantification by qPCR with the KAPA library quantification kit on the ABI7900HT real-time PCR system. This amplification protocol results in 50-200 ng of library in ~26 μl.
The library is now ready for sequencing. If necessary, the library concentration is adjusted to 10 nM by diluting with Elution buffer (see recipes) and submitted for sequencing.
- Amplify ~1 ng of the bisulfite-converted library using a uracil-insensitive PfuTurbo Cx hotstart DNA polymerase and the primer cocktail included in the Illumina TruSeq library construction kit in the following reaction:
Recipes
- Elution buffer
10 mM Tris, pH 8.0
- TE buffer
10 mM Tris-HCl, pH 8.0
1 mM EDTA
Acknowledgments
The protocol was adapted from Wang et al. (2013). This work was supported by a grant from the National Science Foundation (IOS-0842716) awarded to Z.M.
References
- Lister, R., O'Malley, R. C., Tonti-Filippini, J., Gregory, B. D., Berry, C. C., Millar, A. H. and Ecker, J. R. (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133(3): 523-536.
- TruSeq DNA sample preparation guide (PE-940-2001)
(http://nextgen.mgh.harvard.edu/attachments/TruSeq_DNA_SamplePrep_Guide_15005180_A.pdf)
- Wang, Y., An, C., Zhang, X., Yao, J., Zhang, Y., Sun, Y., Yu, F., Amador, D. M. and Mou, Z. (2013). The Arabidopsis Elongator complex subunit2 epigenetically regulates plant immune responses. Plant Cell 25(2): 762-776.
Article Information
Copyright
© 2013 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Amador, D. M., Wang, C., Holland, K. H. and Mou, Z. (2013). Library Construction for Genome-wide Bisulfite Sequencing in Plants. Bio-protocol 3(24): e1013. DOI: 10.21769/BioProtoc.1013.
Category
Plant Science > Plant molecular biology > DNA > DNA sequencing
Molecular Biology > DNA > DNA modification
Systems Biology > Epigenomics > DNA methylation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









