- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Patterned Substrate of Mobile and Immobile Ligands to Probe EphA2 Receptor Clustering
Published: Vol 12, Iss 11, Jun 5, 2022 DOI: 10.21769/BioProtoc.4434 Views: 3040
Reviewed by: David PaulYosuke SenjuJan Huebinger

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Karyopherin-β2 Inhibits and Reverses Aggregation and Liquid-liquid Phase Separation of the ALS/FTD-Associated Protein FUS
Emma Robinson [...] Lin Guo
Aug 20, 2020 5240 Views

Retention Using Selective Hooks (RUSH) Cargo Sorting Assay for Protein Vesicle Tracking in HeLa Cells
Natalia Pacheco-Fernandez [...] Julia von Blume
Mar 5, 2021 9234 Views

In vitro Reconstitution of Phase-separated p62 Bodies on the Arp2/3-derived Actin Network
Tong Liu [...] Na Mi
Apr 20, 2023 2235 Views
Abstract
A multitude of membrane-localized receptors are utilized by cells to integrate both biochemical and physical signals from their microenvironment. The clustering of membrane receptors is widely presumed to have functional consequences for subsequent signal transduction. However, it is experimentally challenging to selectively manipulate receptor clustering without altering other biochemical aspects of the cellular system. Here, we describe a method to fabricate multicomponent, ligand-functionalized microarrays, for spatially segregated and simultaneous monitoring of receptor activation and signaling in individual living cells. While existing micropatterning techniques allow for the display of fixed ligands, this protocol uniquely allows for functionalization of both mobile membrane corrals and immobile polymers with selective ligands, as well as microscopic monitoring of cognate receptor activation at the cell membrane interface. This protocol has been developed to study the effects of clustering on EphA2 signaling transduction. It is potentially applicable to multiple cell signaling systems, or microbe/host interactions.
Graphical abstract:
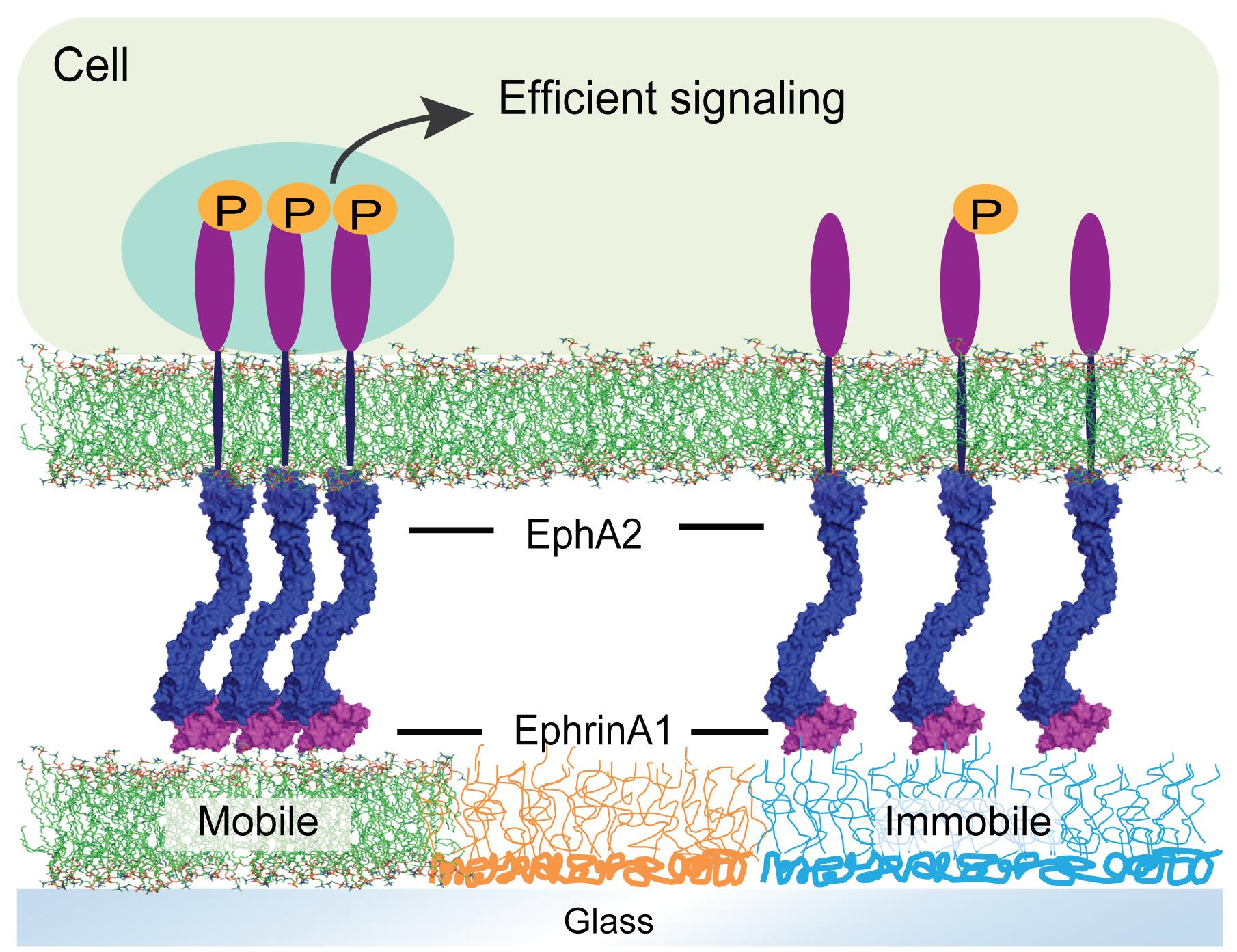
A side-by-side comparison of clustered or non-clustered EphA2 receptor signaling in a single cell.
Background
Cells engage membrane-localized receptors to sense various signals present in their local environment, and respond to these signals appropriately to survive, organize, and proliferate. The signals presented to the cells include both soluble ligands, as well as ligands that are present in the extracellular matrix (ECM), or displayed on the membranes of other cells (also referred to as juxtacrine signaling systems). Membrane receptor interactions of the latter types of ligands enable sensing of the spatial organization of receptor-ligand complexes at the cell-cell interface, and ECM rigidity (Groves and Kuriyan, 2010; Manz and Groves, 2010). A number of these signaling systems have been reconstituted on synthetic supported lipid bilayers in a hybrid format, wherein a live cell interacts with the supported lipid bilayer, recapitulating many of the features of individual receptor types (Biswas and Groves, 2019). These include both reconstitution of the individual receptor signaling system, or a combination of two different receptor signaling systems, to recapitulate the cellular exposure to multiple signals simultaneously, and ensuing signaling crosstalk between the receptors (Chen et al., 2018).
Assembly of cell surface receptors into clusters or organized arrays is a common feature of cell membranes, and has long been implicated as an important factor for modulating signaling activity. However, it is not straightforward to deconvolve the contribution of receptor clustering on signaling itself. A major reason for this is that chemical perturbation of assemblies, such as those achieved with pharmacological agents or mutations (Davis et al., 1994; Seiradake et al., 2013; Bugaj et al., 2013, 2015; Schaupp et al., 2014; Wu et al., 2015; Su et al., 2016), are likely to produce side effects on the cell, other than modulating molecular assembly. Therefore, we seek to modulate spatial organization of receptors by controlling ligand mobility, instead of perturbing intracellular components. In the current protocol, we developed a technique wherein a ligand of interest is displayed in both mobile and immobile configurations, and spatially juxtaposed on length scales small enough to enable a side-by-side comparison within an individual living cell. The immobile ligands are displayed on a functionalized poly L-Lysine-poly ethylene glycol [PLL-(g)-PEG] scaffold, while the mobile ligands are displayed on supported lipid membranes, which allow cluster formation. This method has been applied to the study of EphA2 receptor signaling (Chen et al., 2021), and is potentially applicable to other cell signaling systems or microbe/host interactions (Wong et al., 2021).
Materials and Reagents
Round bottom flask (25 mL)
Glass coverslips (Thorlabs, catalog number: CG15XH)
PLL(20)-g[3.5]-PEG(2)/PEG(3.4)-biotin(50%) (Susos, https://susos.com/shop/pll20-g3-5-peg2peg3-4-biotin50-2/)
PLL(20)-g[3.5]-PEG(3.4)-NTA (Susos, https://susos.com/shop/pll20-g3-5-peg3-4-nta-2/)
PLL(20)-g[3.5]-PEG(2) (Susos, https://susos.com/shop/pll20-g3-5-peg2/)
18:1 (Δ9-Cis) PC (DOPC) (Avanti, catalog number: 850375)
18:1 DGS-NTA(Ni) (Avanti, catalog number:790404)
NeutrAvidin (ThermoFisher, catalog number:22831)
RGD-biotin (Vivitide, catalog number: PCI-3697-PI)
Tris (ThermoFisher, catalog number: 15504020)
NaCl
KCl
CaCl2
MgCl2
D-glucose
EphrinA1 protein (purified in the Groves’ lab, plasmid available upon request)
Alexa FluorTM 680 dye (ThermoFisher, catalog number: A37574)
Chromium-quartz photomask
We use a 5-inch photomask that was fabricated in the Mechanobiology Institute Core facility, at the National University of Singapore. The photomask can also be purchased from other manufacturing companies.
TBS buffer (see Recipes)
Imaging buffer (see Recipes)
Equipment
Imaging chamber (ThermoFisher, AttofluorTM cell imaging chamber, catalog number: A7816)
Water bath
UVO cleaner (Jelight Company INC, hbUVO Cleaner, model 342)
Rotary evaporator (with a standard dry ice condenser, and equipped with a vacuum pump, https://www.asynt.com/product/ika-dry-ice-rotary-evaporator-range/, or https://www.coleparmer.co.uk/i/buchi-23012c000-rotary-evaporator-dry-ice-condenser-220v/2301220)
Tip sonicator (https://www.sonicator.com/collections/sonicators/products/q125-sonicator)
Procedure
This technique requires preparation of (A) small unilamellar vesicles (SUVs), (B) micropatterned polymer surface on a glass substrate, and (C) assembly of membrane arrays on the micropatterned substrate and protein functionalization (Figure 1). The SUVs can be prepared in advance and stored at 4°C for up to 2 weeks. Steps (B) and (C) take 2–3 days, depending on the flexible incubation times.
Briefly, PLL-(g)-PEG scaffold polymers are first coated on a glass coverslip, followed by selective deep UV etching with a photomask and lipid vesicle deposition, to generate regions of immobile polymers and mobile lipid membranes. The ephrinA1 ligands are then functionalized to the substrate in mobile and immobile configurations, to probe EphA2 receptor clustering in cells.
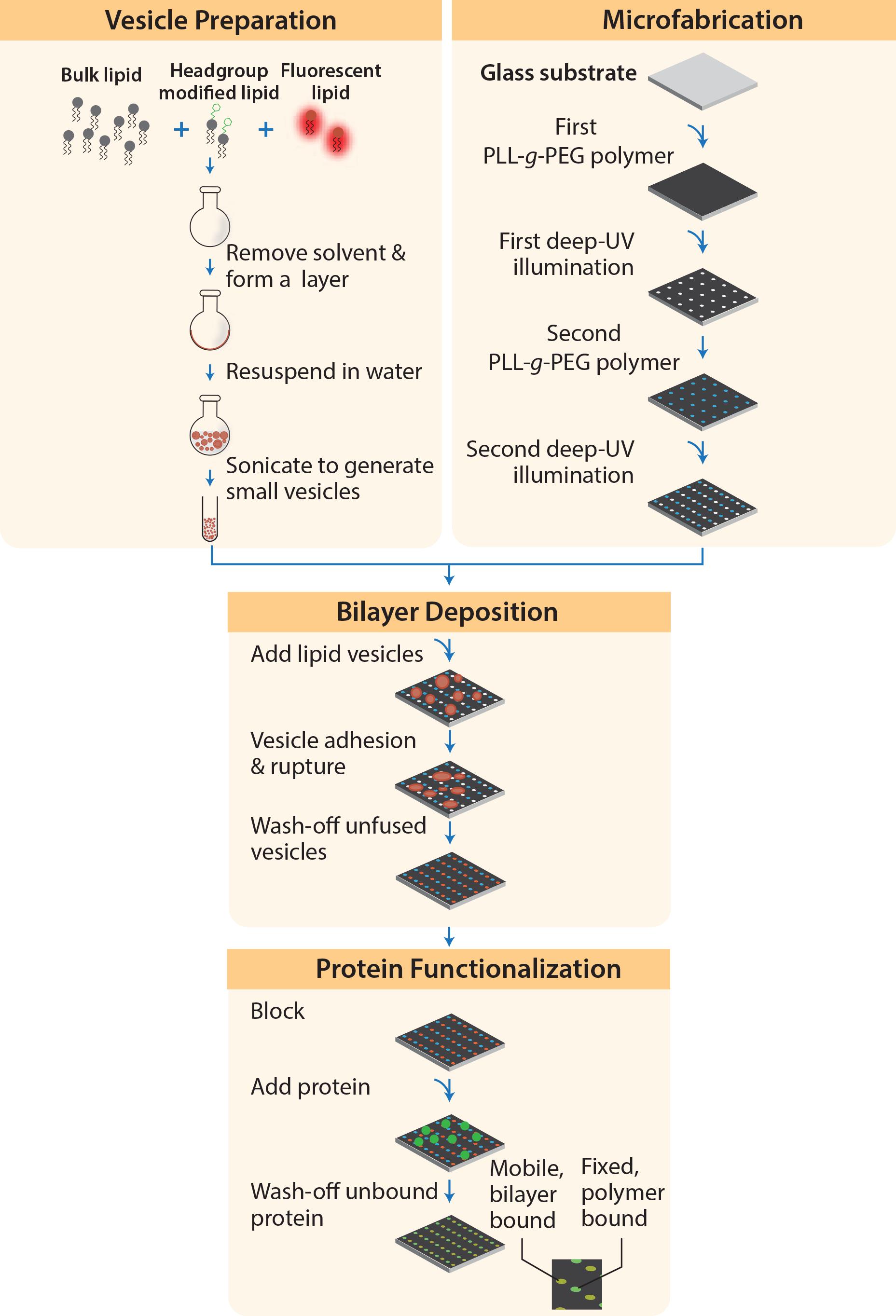
Figure 1. Procedures to Fabricate Micropatterned Substrate of Mobile and Immobile Ligands.
Vesicle preparation
For this step, the audiences can refer to a more detailed protocol (Lin et al., 2009).
Prepare lipid films, by mixing required amounts of various lipid molecules [96% DOPC + 4% DGS-NTA(Ni), with a total mass of 2 mg], which are dissolved in chloroform in a round bottom flask, followed by evaporation of chloroform using a rotary evaporator under vacuum pumping and 50°C water bath, leading to the formation of a thin lipid film in the flask. During this procedure, slowly lower the round bottom flask to the water bath, to avoid chloroform boiling.
Resuspend the lipid film in 2 mL of deionized water by pipetting, allowing the formation of large and often multilamellar vesicles (1 mg/mL).
Sonicate the vesicle suspensions in ice using a probe tip sonicator, to generate SUVs. Typically, use a program of '10 s on, and 5 s off’ for 8–10 cycles for sonication. Transfer the SUV suspension to a fresh tube, and centrifuge at 20,000 × g and 4°C for 2 h, to remove any debris. Transfer the supernatant to a fresh tube, and store at 4°C until further use. The SUVs can be stored for up to 2 weeks, to form a good lipid bilayer.
Microfabrication
Micropatterned surfaces are prepared on glass substrates, using the deep ultraviolet (UV) etching method.
Clean glass coverslips by sonication in a 1:1 mixture of isopropanol and water for 15–30 min, followed by overnight (or longer) incubation in 50% H2SO4 solution. Before usage, take the coverslips out of H2SO4, rinse with water, and expose the glass coverslips to UV light in an enclosed UVO cleaner for 10 min. Rinse with water, and dry by N2 jet.
Incubate the glass coverslips with PLL(20)-g[3.5]-PEG(2)/PEG(3.4)-biotin(50%) at a concentration of 0.1 mg/mL at room temperature for 2 h, or overnight. To reduce the usage of reagents, drop ~30 μL of the incubating solution in parafilm, and lay the glass coverslips on top of it.
Rinse the substrates with water to remove excess polymer, and dry by N2 jet.
Drop ~1.5 μL of water on the micropatterned area (1 cm × 1 cm square) of a chromium-quartz photomask, and lay the polymer-coated glass substrate on its top, to ensure close contact. Expose the substrates to deep UV light for 9–12 min in a UVO cleaner. The UV light-exposed polymer will be degraded. Rinse the substrates with excess water to remove any degraded polymer.
Incubate the same glass substrates with PLL(20)-g[3.5]-PEG(3.4)-NTA at a concentration of 0.1 mg/mL at room temperature for 2 h, and repeat the deep UV etching and washing procedures. After the second etching, the glass substrates should be used for lipid bilayer deposition immediately.
Bilayer deposition and protein functionalization
Before usage, mix SUVs with TBS buffer in 1:1 ratio.
Incubate the micropatterned substrates with SUV solution for 5 min, to allow the self-assembly of lipid bilayers in regions of the substrates that were UV etched in the previous step.
Rinse the substrates with excess TBS buffer, and assemble the glass coverslips into a cell culture imaging chamber, under the aqueous environment.
Block the substrates with bovine serum albumin (BSA) solution (0.05–0.1 mg/mL in TBS buffer) at room temperature for 2h, or at 4°C overnight.
Wash the substrates with TBS buffer, and incubate with NeutrAvidin (1.5 µg/mL) for 15 min, to bind surface biotin groups.
Wash excess NeutrAvidin with TBS buffer, and incubate with ephrinA1-His 10 (purified ephrinA1 protein with a 10-histidine tail in the C-terminus, labeled with Alexa FluorTM 680), and RGD-biotin, for an additional 60–90 min. The ephrinA1-His 10 will bind to both DGS-NTA(Ni) lipids on mobile regions, and PLL(20)-g[3.5]-PEG(3.4)-NTA polymers on immobile regions, while RGD-biotin will bind to NeutrAvidin. The RGD peptides can bind integrins at the cell membrane, to allow cell spreading, during which cells dynamically interact with multiple ephrinA1-functionalized mobile or immobile regions.
Remove unbound proteins by washing with imaging buffer.
Check fluorescence intensity of mobile and immobile ephrinA1 under a microscope.
The substrates are ready for use in a live cell experiment. Prepare cells in the imaging buffer, and seed a low density of cells into the substrate chamber at 37°C for live imaging. The cellular EphA2 receptors will form clusters only after binding with mobile ephrinA1 (Figure 2).
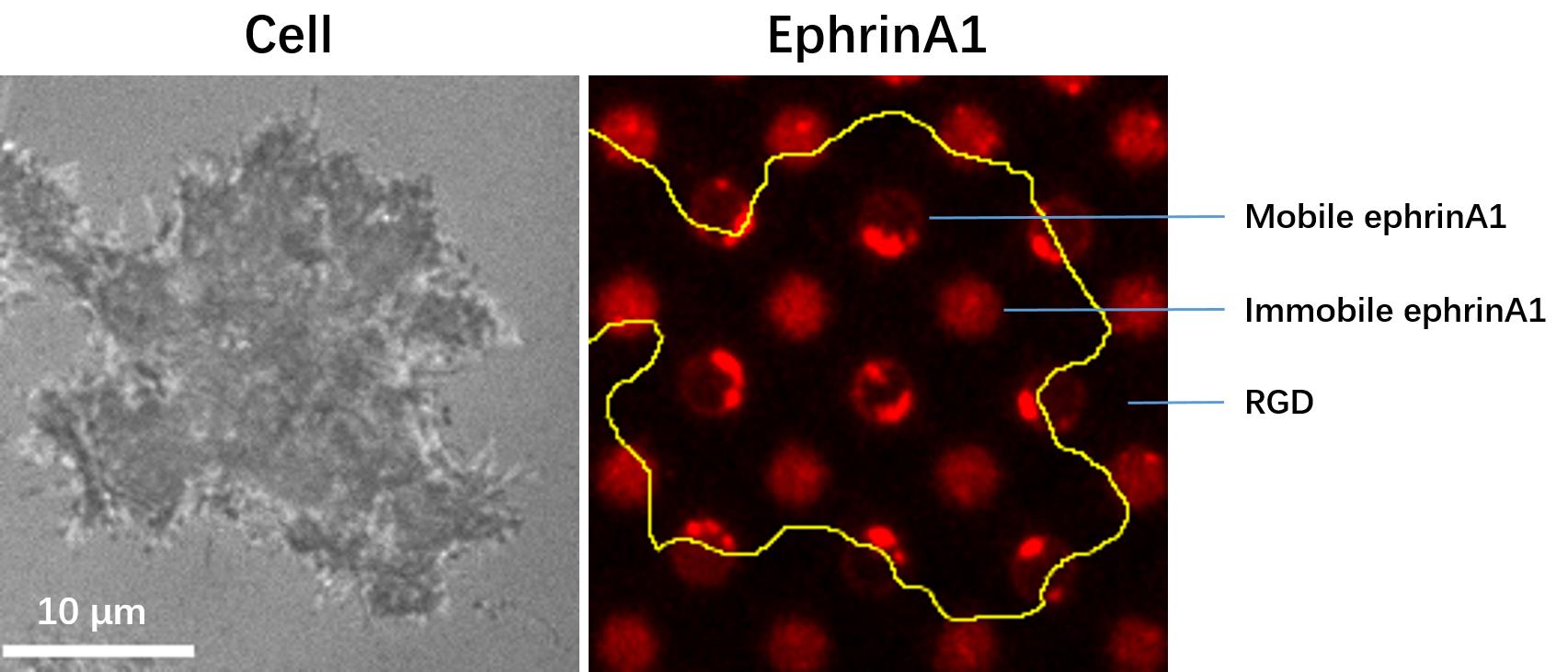
Figure 2. Images of a cell spreading on the micropatterned substrate. Left: The RIC (reflective interference contrast) image showing the cell surface that is adhered to the substrate; Right: The fluorescent image of micropatterned ephrinA1, with the yellow line marking the contour of the cell.
Data analysis
The substrates were observed under a standard fluorescence microscope to compare the fluorescence intensity of mobile and immobile ephrinA1 regions. The mobility of ephrinA1 on lipid bilayers was checked by fluorescence recovery after photobleaching (FRAP).
Notes
The mobile and immobile regions are aligned randomly in two independent UV-etch steps. Usually, the two regions are overlapped in some of the areas, but it is easy to find non-overlapped regions in a centimeter-sized substrate.
The mobile and immobile ephrinA1 intensity may not be the same. Titrate PLL(20)-g[3.5]-PEG(3.4)-NTA with non-reactive PLL(20)-g[3.5]-PEG(2), to modify surface ephrinA1 intensity on immobile regions, or change the lipids molar ratio of DOPC and DGS-NTA(Ni), to modify ephrinA1 intensity on mobile regions.
Recipes
TBS buffer
25 mM Tris
150 mM NaCl
3 mM KCl
Imaging buffer
25 mM Tris
140 mM NaCl
3 mM KCl
2 mM CaCl2
1 mM MgCl2
5.5 mM D-glucose
Acknowledgments
This work was supported by the National Institutes of Health, National Cancer Institute Physical Sciences in Oncology Network Project 1-U01CA202241, and Shanghai Pujiang Program (20PJ1400800). This protocol is derived from published papers (Chen et al., 2018, 2021).
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Biswas, K. H. and Groves, J. T. (2019). Hybrid Live Cell-Supported Membrane Interfaces for Signaling Studies. Annu Rev Biophys 48: 537-562.
- Bugaj, L. J., Choksi, A. T., Mesuda, C. K., Kane, R. S. and Schaffer, D. V. (2013). Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods 10(3): 249-252.
- Bugaj, L. J., Spelke, D. P., Mesuda, C. K., Varedi, M., Kane, R. S. and Schaffer, D. V. (2015). Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nat Commun 6: 6898.
- Chen, Z., Oh, D., Biswas, K. H., Yu, C. H., Zaidel-Bar, R. and Groves, J. T. (2018). Spatially modulated ephrinA1:EphA2 signaling increases local contractility and global focal adhesion dynamics to promote cell motility. Proc Natl Acad Sci U S A 115(25): E5696-E5705.
- Chen, Z., Oh, D., Biswas, K. H., Zaidel-Bar, R. and Groves, J. T. (2021). Probing the effect of clustering on EphA2 receptor signaling efficiency by subcellular control of ligand-receptor mobility. Elife 10: e67379.
- Davis, S., Gale, N. W., Aldrich, T. H., Maisonpierre, P. C., Lhotak, V., Pawson, T., Goldfarb, M. and Yancopoulos, G. D. (1994). Ligands for EPH-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science 266(5186): 816-819.
- Groves, J. T. and Kuriyan, J. (2010). Molecular mechanisms in signal transduction at the membrane. Nat Struct Mol Biol 17(6): 659-665.
- Lin, W. C., Yu, C. H., Triffo, S. and Groves, J. T. (2010). Supported membrane formation, characterization, functionalization, and patterning for application in biological science and technology. Curr Protoc Chem Biol 2(4): 235-269.
- Manz, B. N. and Groves, J. T. (2010). Spatial organization and signal transduction at intercellular junctions. Nat Rev Mol Cell Biol 11(5): 342-352.
- Schaupp, A., Sabet, O., Dudanova, I., Ponserre, M., Bastiaens, P. and Klein, R. (2014). The composition of EphB2 clusters determines the strength in the cellular repulsion response. J Cell Biol 204(3): 409-422.
- Seiradake, E., Schaupp, A., del Toro Ruiz, D., Kaufmann, R., Mitakidis, N., Harlos, K., Aricescu, A. R., Klein, R. and Jones, E. Y. (2013). Structurally encoded intraclass differences in EphA clusters drive distinct cell responses. Nat Struct Mol Biol 20(8): 958-964.
- Su, X., Ditlev, J. A., Hui, E., Xing, W., Banjade, S., Okrut, J., King, D. S., Taunton, J., Rosen, M. K. and Vale, R. D. (2016). Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352(6285): 595-599.
- Wong, J. J., Chen, Z., Chung, J. K., Groves, J. T. and Jardetzky, T. S. (2021). EphrinB2 clustering by Nipah virus G is required to activate and trap F intermediates at supported lipid bilayer-cell interfaces. Sci Adv 7(5): eabe1235.
- Wu, Y., Kanchanawong, P. and Zaidel-Bar, R. (2015). Actin-delimited adhesion-independent clustering of E-cadherin forms the nanoscale building blocks of adherens junctions. Dev Cell 32(2): 139-154.
Article Information
Copyright
Chen et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Chen, Z., Biswas, K. H. and Groves, J. T. (2022). Patterned Substrate of Mobile and Immobile Ligands to Probe EphA2 Receptor Clustering. Bio-protocol 12(11): e4434. DOI: 10.21769/BioProtoc.4434.
- Chen, Z., Oh, D., Biswas, K. H., Zaidel-Bar, R. and Groves, J. T. (2021). Probing the effect of clustering on EphA2 receptor signaling efficiency by subcellular control of ligand-receptor mobility. Elife 10: e67379.
Category
Biological Engineering > Synthetic biology
Biophysics > Bioengineering
Biochemistry > Protein > Self-assembly
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









