- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Nitrate Reductase Activity Assay of Mycolicibacterium smegmatis Crude Extract
Published: Vol 11, Iss 14, Jul 20, 2021 DOI: 10.21769/BioProtoc.4098 Views: 3298
Reviewed by: Partha Basu Valentine V Trotter

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

H2 Production from Methyl Viologen–Dependent Hydrogenase Activity Monitored by Gas Chromatography
Nuttavut Kosem
Dec 5, 2023 1758 Views

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1575 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1916 Views
Abstract
Nitrate is one of the major inorganic nitrogen sources for microorganisms. Many bacterial and archaeal lineages can express cytoplasmic assimilatory nitrate reductase (NAS), which catalyzes the rate-limiting reduction of nitrate to nitrite in the nitrate assimilation pathway. Here, we present a detailed protocol for measuring in vitro nitrate reductase (NaR) activity of NAS enzymes from Mycolicibacterium smegmatis crude extract using both physiological and non-physiological electron donors.
Keywords: Mycolicibacterium smegmatisBackground
In addition to being an important nitrogen source for plants, nitrate (NO3-) is universally used as an inorganic nitrogen source for microorganisms, particularly soil bacteria such as actinomycetes, as well as marine bacteria such as cyanobacteria, due to fierce competition for limited nutrients in the natural environment (Kuypers et al., 2018). In prokaryotes, nitrate is reduced to ammonia (NH3) through two sequential steps, with the first committed step catalyzed by NAS. Prokaryotic NASs are categorized based on the physiological electron donors utilized (Morozkina and Zvyagilskaya, 2007; Tan et al., 2020), i.e., the flavodoxin- or ferredoxin-dependent NAS and the NAD(P)H-dependent NAS; however, to date, there have been few reports detecting in vitro NaR activity of bacterial NAD(P)H-dependent NASs employing their physiological electron donors. Instead, methyl viologen (an artificial chemical reductant) is the most commonly used electron donor for in vitro NaR activity assays.
In this protocol, employing an optimized assay system, NAD(P)H-dependent NaR activity can be measured in M. smegmatis crude cell extract. Briefly, the enzyme samples prepared under anaerobic conditions can use both NADPH and NADH as electron donors for catalysis in vitro. Nitrite produced by nitrate reduction forms a colored azo compound with sulfanilamide and N-(1-naphthyl)ethylenediamine dihydrochloride, which can be measured by its absorbance at 540 nm. This method is suitable for the determination of in vitro NaR activity of bacterial NAS enzymes.
Materials and Reagents
Mycolicibacterium smegmatis mc2155
Middlebrook 7H10 agar (BD Biosciences, catalog number: 262710)
Middlebrook 7H9 broth (BD Biosciences, catalog number: 271310)
Glycerol (Sigma-Aldrich, catalog number: G5516)
Tween 80 (Sigma-Aldrich, catalog number: P1754)
Ethanol (Sigma-Aldrich, catalog number: 51976)
Phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626)
Bradford protein assay kit (Beyotime, catalog number: P0006)
Sodium nitrate (NaNO3) (Sigma-Aldrich, catalog number: S5506)
Sodium nitrite (NaNO2) (Sigma-Aldrich, catalog number: S2252)
Sodium phosphate dibasic (Na2HPO4) (Sigma-Aldrich, catalog number: 255793)
Sodium phosphate monobasic (NaH2PO4) (Sigma-Aldrich, catalog number: S3139)
Nicotinamide adenine dinucleotide phosphate (NADPH) (Sigma-Aldrich, catalog number: N7505)
Nicotinamide adenine dinucleotide (NADH) (Sigma-Aldrich, catalog number: 43420)
Methyl viologen (Sigma-Aldrich, catalog number: 856177)
Flavin mononucleotide (FMN) (Sigma-Aldrich, catalog number: F2253)
Flavin adenine dinucleotide (FAD) (Sigma-Aldrich, catalog number: F6625)
Sodium hydrosulfite (Na2S2O4) (Sigma-Aldrich, catalog number: 71699)
Sodium bicarbonate (NaHCO3) (Sigma-Aldrich, catalog number: S6014)
Sulfanilamide (Sigma-Aldrich, catalog number: S9251)
N-(1-naphthyl)ethylenediamine dihydrochloride (NED) (Sigma-Aldrich, catalog number: 222488)
7H10 agar (see Recipes)
7H9 medium (see Recipes)
MPLN medium (see Recipes)
0.5 M sodium phosphate buffer, pH 7.5 (see Recipes)
0.1 M sodium phosphate buffer, pH 7.5 (see Recipes)
0.1 M PMSF (see Recipes)
60 mM Na2S2O4/NaHCO3 (see Recipes)
1% (w/v) sulfanilamide solution (see Recipes)
0.01% (w/v) NED solution (see Recipes)
Equipment
Pipette kit (Gilson, catalog number: F167300)
High-speed centrifuge (Beckman, catalog number: AVANTI JXN-26)
Ultrasonic cell disruptor (Beyotime, catalog number: E0385)
Ultrasonic bath equipped with a degas mode (Beyotime, catalog number: E0439)
250-ml and 2-L Erlenmeyer flasks
Petri dish (90 mm diameter)
37°C incubator
Incubator shaker
Water bath
Spectrophotometer
Anaerobic chamber (glove box) filled with mixed gas (nitrogen, 95%; hydrogen, 5%)
Note: Place the pipette kit, ultrasonic cell disruptor, and water bath inside the anaerobic chamber.
Software
Microsoft Excel
Procedure
Preparation of M. smegmatis culture
Streak M. smegmatis frozen culture on a 7H10 agar plate and incubate aerobically at 37°C for 3 days until colonies are visible.
Inoculate a single colony into 50 ml 7H9 medium in a 250-ml Erlenmeyer flask and incubate aerobically at 37°C for 16 h with shaking at 180 rpm (overnight culture).
Collect cells from the overnight culture by centrifuging at 3,000 × g for 10 min. Wash the pellet twice with nitrogen-free MPLN medium and centrifuge as above. Resuspend the cells in 50 ml MPLN medium as the seeding culture.
Re-inoculate 10 ml seeding culture into 1 L MPLN medium supplemented with 10 mM nitrate in a 2-L Erlenmeyer flask and incubate aerobically at 37°C for 30 h with shaking to late-log phase (OD600 0.8–1.0).
Preparation of M. smegmatis crude extract
Collect cells by centrifuging at 5,000 × g for 10 min at 4°C.
Wash the cell pellet twice with 30 ml degassed 0.1 M sodium phosphate buffer (pH 7.5).
Transfer the cell pellet into an anaerobic chamber and resuspend in 20 ml degassed 0.1 M sodium phosphate buffer (pH 7.5) supplemented with 10% v/v glycerol and 1 mM PMSF.
Disrupt cells by sonication (pulse: 0.3 s, stop: 0.7 s) for 10 min on ice and transfer the lysate to a centrifuge tube inside the anaerobic chamber.
Centrifuge at 16,000 × g for 1 h at 4°C outside the anaerobic chamber.
Bring the centrifuge tube back into the anaerobic chamber and collect the supernatant (cytoplasmic fraction) as the crude enzyme extract.
Note: Keep on ice during the assay and use within 24 h.
Determine the total protein concentration of the cytoplasmic fraction using the Bradford method.
Preparation of the nitrite standard curve
Prepare a nitrite dilution series using sodium nitrite (0, 3, 30, 60, 120, 240, and 300 μM) in deionized water.
Pipette 0.5 ml nitrite dilution series (Test sample) into 1.5-ml tubes. Use 0.5 ml 10 mM sodium nitrate as a control (Blank sample).
Add 0.5 ml sulfanilamide solution and 0.5 ml NED solution to all tubes.
Mix by swirling and incubate at 25°C for 10 min.
Transfer 1 ml assay mixture to a cuvette.
Measure the visible absorption at 540 nm using the spectrophotometer.
Calculate the absorbance change using the following equation:
ΔA540 nm (Sample) = A540 nm (Test) - A540 nm (Blank)Plot the standard curve with the nitrite concentration on the horizontal axis and the absorbance change on the vertical axis (Figure 1).
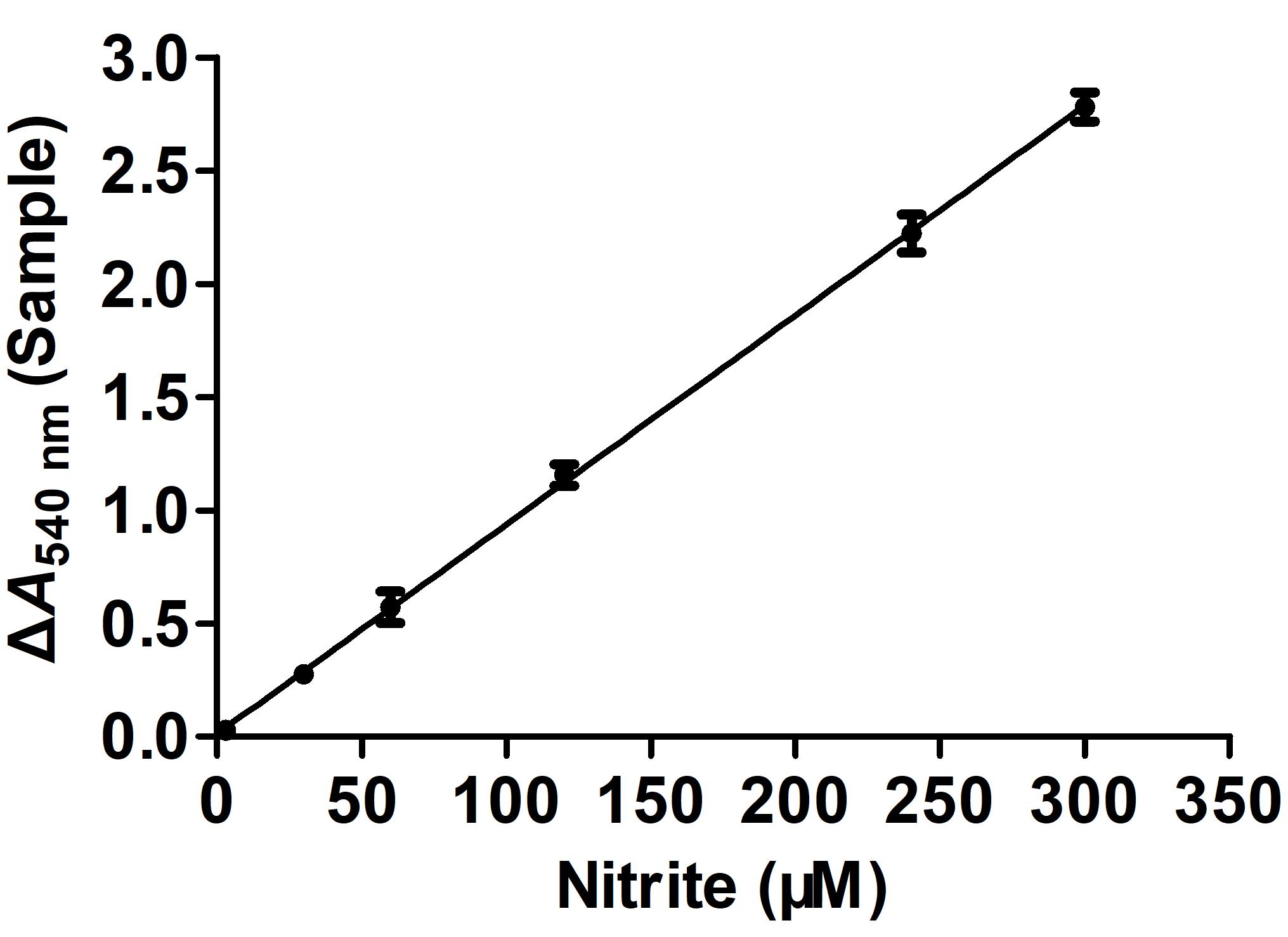
Figure 1. Nitrite standard curve. Error bars indicate the average value of three biological replicates ± SD (n = 3). According to the standard curve, the regression equation is x (µM) = (y-0.0204)/0.0092, where x stands for nitrite concentration and y stands for ΔA540 nm (Sample).
In vitro NaR activity assay
Before preparing the reaction mixture for the enzyme assay, degas the solutions used in the assays and place in the anaerobic chamber before use.
Pipette the following reagents for the nitrate reduction reaction (Tables 1 and 2) into a 1.5-ml tube inside the anaerobic chamber:
Table 1. Assay mixtures for determining NAD(P)H-dependent NaR activity
Assay sample Test Blank Deionized water 0.090 ml 0.100 ml Sodium phosphate buffer (0.5 M) 0.100 ml 0.100 ml NaNO3 (50 mM) 0.100 ml 0.100 ml NADPH (2 mM) or NADH (2 mM) 0.100 ml 0.100 ml FMN (0.25 mM) 0.050 ml 0.050 ml FAD (0.25 mM) 0.050 ml 0.050 ml Crude enzyme extract 0.010 ml - Step 1: Mix gently and incubate at 30°C for 10 min inside the anaerobic chamber 1% (w/v) sulfanilamide solution 0.500 ml 0.500 ml 0.01% (w/v) NED solution 0.500 ml 0.500 ml Step 2: Mix gently and incubate at 25°C in the dark for 10 min inside the anaerobic chamber Table 2. Assay mixtures for determining methyl viologen-dependent NaR activity
Assay sample Test Blank Deionized water 0.090 ml 0.100 ml Sodium phosphate buffer (0.5 M) 0.100 ml 0.100 ml NaNO3 (50 mM) 0.100 ml 0.100 ml Methyl viologen (0.75 mM) 0.100 ml 0.100 ml Na2S2O4/NaHCO3 (60 mM) 0.100 ml 0.100 ml Crude enzyme extract 0.010 ml - Step 1: Mix gently and incubate at 30°C for 10 min inside the anaerobic chamber
Step 2: Stop the reaction by vigorous stirring in the open air outside the anaerobic chamber until the blue color disappears1% (w/v) sulfanilamide solution 0.500 ml 0.500 ml 0.01% (w/v) NED solution 0.500 ml 0.500 ml Step 3: Mix gently and incubate at 25°C in the dark for 10 min outside the anaerobic chamber Note: The assay mixture turns deep blue after adding Na2S2O4/NaHCO3, an indication of methyl viologen being reduced.
Transfer 1 ml assay mixture to a cuvette.
Measure the visible absorption at 540 nm for both the Test and Blank samples.
Calculate the NaR specific activity using the following equation:
NaR specific activity (nmol/min/mg) = Cnitrite DF/T/Cenzyme/Venzyme
where,
Cnitrite (µM) = [A540 nm (Test) - A540 nm (Blank)-0.0204]/0.0092, the concentration of nitrite formed in the assay
DF: The dilution factor
T (min): The reaction time for nitrate reduction in the assay
Note: An incubation time of 10 min is sufficient for measuring the in vitro nitrate reductase activity.
Cenzyme (mg/ml): The protein concentration of the crude enzyme extract
Venzyme (ml): The volume of the crude enzyme extract used in the assay
Recipes
7H10 agar
Dissolve 19 g 7H10 powder in 1 L deionized water
Add 4 ml 50% (v/v) glycerol and 5 ml 10% (v/v) Tween 80
Autoclave at 121°C for 15 min
7H9 medium
Dissolve 4.7 g 7H9 powder in 1 L deionized water
Add 4 ml 50% (v/v) glycerol and 5 ml 10% (v/v) Tween 80
Autoclave at 121°C for 15 min
Nitrogen-free MPLN medium (pH 6.6)
36.7 mM KH2PO4
7.7 mM sodium citrate
5 mM MgSO4
60 µM FeCl3
20 µM NaMoO4
3.4 µM CaCl2
3.5 µM ZnSO4
4 µM CuSO4
2% (v/v) glycerol
0.05% (v/v) Tween 80
Autoclave at 121°C for 15 min.
0.5 M sodium phosphate buffer (pH 7.50)
Prepare 0.5 M solutions of NaH2PO4 and Na2HPO4
Mix 20% (v/v) NaH2PO4 and 80% (v/v) Na2HPO4
Adjust pH to 7.5
0.1 M sodium phosphate buffer (pH 7.50)
Dilute 0.5 M sodium phosphate buffer 1:5 with deionized water
Store at room temperature
0.1 M PMSF
Dissolve 0.174 g PMSF in 10 ml ethanol
Store at -20°C
60 mM Na2S2O4/NaHCO3
Prepare 120 mM solutions of Na2S2O4 and NaHCO3
Mix 50% (v/v) Na2S2O4 and 50% (v/v) NaHCO3
Note: Prepare fresh
1% (w/v) sulfanilamide solution
Dissolve in 3 M HCl
Note: Prepare fresh and store at 4°C in the dark
0.01% (w/v) NED solution
Dissolve in deionized water
Note: Prepare fresh and store at 4°C in the dark
Acknowledgments
This protocol was adapted from a previously published paper by Tan et al. (2020). This research was supported by the National Natural Science Foundation of China (31430004, 31830002, 81671988, 91751000, 91951000 to G.-P. Z.), the Research Unit Fund of Li Ka Shing Institute of Health Sciences (7103506 to G.-P. Z.), and the Hong Kong Health and Medical Research Fund (12110622 to G.-P. Z.).
Competing interests
The authors declare that they have no conflicts of interest.
References
- Kuypers, M. M. M., Marchant, H. K., and Kartal, B. (2018). The microbial nitrogen-cycling network. Nat Rev Microbiol 16(5): 263-276.
- Morozkina, E. V., and Zvyagilskaya, R. A. (2007). Nitrate reductases: structure, functions, and effect of stress factors. Biochemistry (Mosc) 72(10): 1151-1160.
- Tan, W., Liao, T. H., Wang, J., Ye, Y., Wei, Y. C., Zhou, H. K., Xiao, Y., Zhi, X. Y., Shao, Z. H., Lyu, L. D. and Zhao, G. P. (2020). A recently evolved diflavin-containing monomeric nitrate reductase is responsible for highly efficient bacterial nitrate assimilation. J Biol Chem 295(15): 5051-5066
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Tan, W., Shao, Z. and Zhao, G. (2021). In vitro Nitrate Reductase Activity Assay of Mycolicibacterium smegmatis Crude Extract. Bio-protocol 11(14): e4098. DOI: 10.21769/BioProtoc.4098.
- Tan, W., Liao, T. H., Wang, J., Ye, Y., Wei, Y. C., Zhou, H. K., Xiao, Y., Zhi, X. Y., Shao, Z. H., Lyu, L. D. and Zhao, G. P. (2020). A recently evolved diflavin-containing monomeric nitrate reductase is responsible for highly efficient bacterial nitrate assimilation. J Biol Chem 295(15): 5051-5066
Category
Microbiology > Microbial biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









