- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Reconstitution Assays of Arabidopsis 20S Proteasome
Published: Vol 11, Iss 7, Apr 5, 2021 DOI: 10.21769/BioProtoc.3967 Views: 5776
Reviewed by: Wenrong HeYuan WangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2855 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1975 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1763 Views
Abstract
The majority of cellular proteins are degraded by the 26S proteasome in eukaryotes. However, intrinsically disordered proteins (IDPs), which contain large portions of unstructured regions and are inherently unstable, are degraded via the ubiquitin-independent 20S proteasome. Emerging evidence indicates that plant IDP homeostasis may also be controlled by the 20S proteasome. Relatively little is known about the specific functions of the 20S proteasome and the regulatory mechanisms of IDP degradation in plants compared to other species because there is a lack of systematic protocols for in vitro assembly of this complex to perform in vitro degradation assays. Here, we present a detailed protocol of in vitro reconstitution assay of the 20S proteasome in Arabidopsis by modifying previously reported methods. The main strategy to obtain the 20S core proteasome here is to strip away the 19S regulatory subunits from the 26S proteasome. The protocol has two major parts: 1) Affinity purification of 20S proteasomes from stable transgenic lines expressing epitope-tagged PAG1, an essential component of the 20S proteasome (Procedures A-D) and 2) an in vitro 20S proteasome degradation assay (Procedure E). We anticipate that these protocols will provide simple and effective approaches to study in vitro degradation by the 20S proteasome and advance the study of protein metabolism in plants.
Keywords: 26S proteasomeBackground
In eukaryotes, protein degradation is carried out by the proteasome. The integrative 26S proteasome is comprised of two sub particles: one or two terminal 19S regulatory particle(s) (RP), which serve as a proteasome activator; and the 20S core proteasome (CP), which degrades proteins. Most eukaryotic proteins are polyubiquitinated and channeled into 26S proteasome for degradation. In contrast, proteins that contain intrinsically disordered regions have been found to be directly destroyed by the ubiquitin-independent 20S proteasome (Ben-Nissan et al., 2014). Methods to purify and assembly the 20S proteasome in vitro are well established for mammalian cells and yeast. This has led to an understanding and appreciation of the numerous ways by which IDPs interact with the 20S proteasome (Leggett et al., 2005). However, to date a detailed and efficient protocol has not been reported for the purification of the 20S proteasome from plants. Book et al. (2010) developed an affinity-based strategy to effectively isolate the 26S proteasome from Arabidopsis. In their approach, PAG1, one of the 14 core proteasome polypeptides, is epitope-tagged and immunoprecipitated with epitope-specific antibodies such that the 26S proteasome is recovered. From purification studies of the proteasome complex, there are two important components to monitor during the purification scheme: ATP amount and salt concentration (Verma et al., 2000; Leggett et al., 2002). It is known that the integrity of the RP-CP complex relies on ATP, and the abundance of RP subunits is substantially reduced if all purification steps do not include ATP. Similarly, RP subunit abundance is reduced when immunoprecipitates (IPs) are washed with a high salt buffer (800 mM NaCl) before elution (Book et al., 2010). Based on previous methods, we designed a simple approach to specifically isolate the 20S proteasome by immunoprecipitating PAG1 complexes from total protein extracts of stable transgenic lines expressing gPAG1-Flag-4Myc (gPAG1-FM) under its native promoter. In our protocol, the protein extracts are not supplied with ATP, and IPs are washed with a buffer containing 800 mM NaCl, as this stringent condition has been reported to strip the 19S regulatory subunits away from the 20S core proteasome. For in vitro protein degradation assays, we modified a protocol from yeast work with the 20S proteasome (Hsieh et al., 2015). Taken together, this protocol is easy to follow and can provide an effective strategy to study degradation of IDPs in plants. We hope that this protocol will advance research in protein metabolism and regulation in plants.
Materials and Reagents
15 ml conical tube
2 ml Eppendorf tube
1.5 ml Eppendorf tube
Pipette tips
96-well Plate (Thermo Scientific, catalog number: 249935 )
10-day-old PPAG1-gPAG1-FM transgenic seedlings
Liquid nitrogen
Tris Base (Fisher Scientific, catalog number: BP152-10 )
Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771 )
NaCl (Fisher Scientific, catalog number: BP358-10 )
MgCl2 (Sigma-Aldrich, catalog number: M9272 )
EDTA (Fisher Scientific, catalog number: BP120-1 )
Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43817 )
Glycerol (Sigma-Aldrich, catalog number: V900122 )
PMSF (Sigma-Aldrich, catalog number: 78830 )
Miracloth (Calbiochem, catalog number: 475855 )
Anti-FLAG® M2 magnetic beads (Sigma-Aldrich, catalog number: M8823 )
3× FLAG peptide (DYKDDDDK) (Sigma-Aldrich, catalog number: F4799 )
Anti-Flag (Sigma-Aldrich, catalog number: F1804 )
Anti-Myc (Sigma-Aldrich, catalog number: C3956 )
Anti-SE (Agrisera, catalog number: AS09 532A)
DMSO (Sigma-Aldrich, catalog number: D4540 )
MG132 (Calbiochem, catalog number: 474787 )
Succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (Suc-LLVY-AMC) (Sigma-Aldrich, catalog number: S6510 )
SilverQuestTM Staining Kit (Invitrogen, catalog number: LC6070 )
Bradford Reagent (Sigma-Aldrich, catalog number: B6916 )
2× SDS-PAGE loading buffer (see Recipes)
Extraction buffer (see Recipes)
Washing buffer (see Recipes)
Reaction Buffer (see Recipes)
Equipment
-80 °C freezer
Pipettes
Centrifuge
DynaMagTM-2 (Invitrogen, model: 12321D )
PolyATtract® System 1000 Magnetic Separation Stand (Promega Corporation, model: Z5410 )
Rugged Rotator (Glas-Col, model: 099A RD4512 )
Thermomixer R (Eppendorf, model: 022679810)
Microplate spectrophotometer (PerkinElmer, model: VICTORTM X3)
Gel imaging system (Bio-Rad, model: Universal Hood II )
Vortex mixer (VWR, model: 945300 )
Procedure
Note: Please see Figure 1 for a schematic diagram of the steps described below.

Figure 1. A schematic diagram of procedure
Preparation of 10-day-old PPAG1-gPAG1-FM stable transgenic plants (Li et al., 2020)
Transform a binary vector pBA002a-PPAG1-gPAG1-FM into Col-0 ecotype of Arabidposis thaliana by the floral-dip transformation method (Zhang et al., 2006) to generate PPAG1-gPAG1-FM stable transgenic plants.
Sterilize and place the seeds from the stable transgenic line expressing PPAG1-gPAG1-FM on MS medium (Zhang et al., 2006) and stratify seeds by keeping them in the dark at 4 °C for 3 days.
Germinate seeds and grow the seedlings under a 12 h light-12 h dark cycle at 22 °C for 10 days.
Collect 5 g of 10-day-old seedlings, ground to fine powder in liquid nitrogen, and stored at -80 °C.
Affinity purification of 20S proteasomes (Book et al., 2010)
Re-suspend the 5 g powder sample in 8 ml of extraction buffer.
Keep the sample on ice for min. Keeping the sample cold, homogenize it well with a vortex mixer 2-3 times.
Centrifuge the fully dissolved protein extract at 4 °C for 15 min at 21,000 × g and then filter the protein extract through one layer of pre-wet Miracloth.
After filtration, centrifuge the cleared protein extract again at 4 °C for 15 min at 21,000 × g.
Collect the supernatant from Step B4 in a pre-cooled 15 ml conical tube and keep it on ice.
Prepare Anti-FLAG beads for immunoprecipitation during the centrifugation steps. Add 200 μl bed volume Anti-FLAG® M2 magnetic beads (for 5 g plant tissue) into a new 2 ml Eppendorf tube on ice.
Wash the magnetic FLAG-beads with 600 μl of 0.1 M glycine HCl (pH 3.0) to remove un-conjugated antibody.
Invert the tube gently and leave the mixture in the tube for 1.5 min.
Immediately re-equilibrate the Anti-FLAG® M2 magnetic beads with 1 ml buffer (50 mM Tris-HCl, 150 mM NaCl, pH 8.0).
Remove the equilibration buffer, followed by washing the beads with 1 ml extraction buffer for three times using DynaMag10. Remove the equilibration buffer, followed by washing the beads with 1 ml extraction buffer three times using DynaMagTM-2.
Completely remove the extraction buffer and add 200 μl new extraction buffer into the tube to re-suspend the beads.
Add 200 μl equilibrated magnetic beads into sample.
Rotate the 15 ml tube at 4 °C for 30 min using a rugged rotator.
At the end of Step B13, prepare 3× FLAG peptide solution for elution of Flag-4Myc-tagged PAG1 from the Anti-FLAG® M2 magnetic beads.
Add 35 μl 3× FLAG elution buffer stock (4 mg/ml) into 245 μl extraction buffer to make a final concentration 500 ng/μl of 3× FLAG peptide. Mix well and put it on ice.
After Step B13 is done, load the 15 ml tube on the PolyATtract® System 1000 Magnetic Separation Stand for a few seconds, then slowly pour out the supernatant. Supernatant can be discarded.
Re-suspend the beads with 6 ml washing buffer and transfer all the beads to a new, clean precooled 15 ml tube.
Wash the beads with 6 ml washing buffer three times at 4 °C, each time for 5 min using the rugged rotator.
After washing three times, add 2 ml of extraction buffer to a 15 ml tube to re-suspend the beads.
Transfer all the beads carefully to a clean 2 ml Eppendorf tube.
Load the 2 ml tube into DynaMagTM-2, and remove the extraction buffer.
Add 250 μl 3× FLAG elution buffer into the 2 ml tube, incubate the tube for 30 min at 4°C with 1,200 rpm shaking using Thermomixer R.
Load the 2 ml tube into DynaMagTM-2. Transfer the eluates to the 1.5 ml Eppendorf tube and store at -80 °C.
Silver staining of purified proteasome
Add 20 μl 2× SDS-PAGE loading buffer into 20 μl of the eluted sample and boil at 95 °C for 8 min.
Load 20 μl eluted sample onto two 10% SDS-PAGE gels and subject to electrophoresis for 2.5 h at 80 V.
Stain one of the gels with SilverQuestTM Staining Kit (Figure 2).
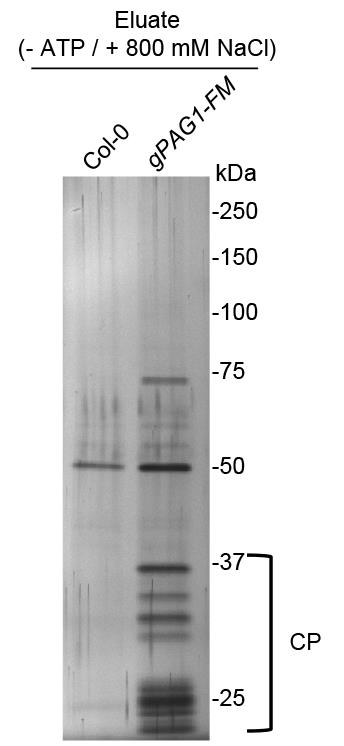
Figure 2. A representative silver-staining image of immunoprecipitated PAG1-FM-containing 20S resolved by SDS-PAGE. The immunoprecipitation was performed using the anti-FLAG® M2 magnetic beads with the protein extracts prepared from PPAG1-gPAG1-FM transgenic seedlings (gPAG1-FM) or from Col-0 (control), respectively. The bracket indicates subunits of the 20S core proteasome (CP).Do Western blot analysis for the other gel using anti-Myc or anti-Flag antibodies.
Take images of silver staining gel using a gel imaging documentation system.
Proteasome activity assay (Yang et al., 2004; Han et al., 2019)
Prepare reaction buffer containing 50 μM Suc-LLVY-AMC substrate, which is widely used as a fluorogenic substrate for measuring the chymotrypsin – like activity of the 20S proteasome (Reidlinger et al., 1997).
Add 10 μl eluted sample into 90 μl reaction Buffer, mix well and add into 96-well Plate.
Add 10 μl extraction buffer into 90 μl reaction Buffer as a blank control and repeat each reaction three times.
Incubate the reaction mixtures at 37 °C for 20, 40, 60, 80, 100, 120 min.
Monitor the fluorescence reading of the released AMC at the indicated times using a Microplate spectrophotometer by fluorescence using 380 nm excitation and 440 nm emission wavelengths.
Plot proteasome activity in relative fluorescence units per 1 μl of reaction mixtures, using free AMC as a standard.
Note: You can also use other representations to display your data, such as relative fluorescence units per 10 μl of eluted sample.
In vitro 20S proteasome degradation assay (Hsieh et al., 2015)
Prepare purified proteins for testing.
Estimate the concentration of the purified proteasome and test proteins by the Bradford method (Bradford, 1976). (Method: https://www.sigmaaldrich.com/content/dam/sigma-aldrich/docs/Sigma/Bulletin/b6916bul.pdf).
Prepare in vitro 20S proteasome-decay reaction mixtures on ice as follows (Table 1):
Table 1. In vitro 20S proteasome degradation reaction system
Note: The amount of the purified 20S proteasome and test protein in the reaction mixture is only a reference, the reaction condition needs to be optimized for different proteins.One reaction Reaction mixtures for five indicated times 150 nM Purified test protein According to the concentration 10 nM Purified 20S proteasome According to the concentration 50 mM Tris-HCl (pH 7.5) 10 μl [1 M Tris-HCl (pH 7.5) ] 2% DMSO or 50 μM MG132 4 μl (2.5 mM MG132, dissolved in DMSO) Add H2O to final volume 40 μl Add H2O to final volume 200 μl Then distribute the mixtures evenly into five PCR or 1.5 ml tubes and incubate the tubes at 22 °C.
Stop the reaction by adding 40 μl 2× SDS-PAGE loading buffer at the indicated times (0, 5, 10, 20, 30 min) followed by Western blot analysis (Figure 3).
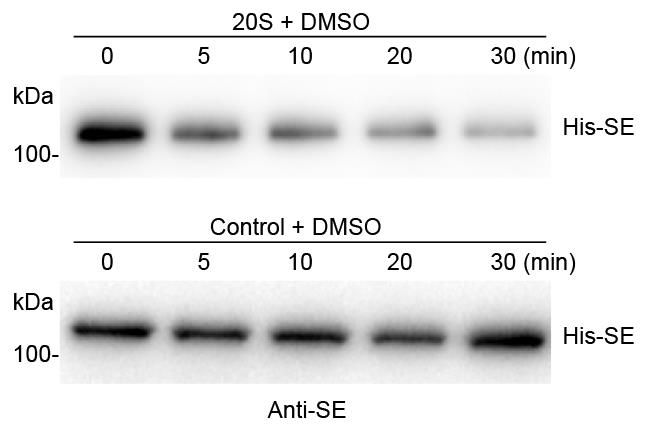
Figure 3. A representative image of in vitro 20S proteasome-mediated protein degradation assay. Recombinant 6xHis-SUMO-SE protein was incubated with the PAG1-FM immunoprecipitate from PPAG1-gPAG1-FM transgenic plants or control IP from Col-0, respectively. The reaction mixture was stopped at the indicated time intervals. Western blot assay of 6xHis-SE was probed with an anti-SE antibody.
Data analysis
For additional reference images of silver staining and in vitro 20S proteasome degradation assay, see Figures 4c and 4d from Li et al. (2020), respectively. For additional reference images of the proteasome activity assay, see the Extended Data Figure 7b from Li et al. (2020).
Notes
In step B, all the buffers and tubes required for purification should be pre-cooled to 4 °C before use.
In step B, Miracloth is pre-wet with extraction buffer.
In step B, divide the final elution samples into several tubes to avoid freeze-thaw samples in future use, which will effect proteasome activity.
In step E, optimize the in vitro degradation conditions according to test proteins, including pH, degradation time, degradation temperature and the concentration of the 20S proteasome and test proteins.
In step E, prepare and evenly distribute the reaction mixtures quickly and keep on ice. Make sure all time points start the reaction at the same time.
Recipes
2× SDS-PAGE loading buffer
0.125 mM Tris-HCl, pH 6.8
20% glycerol
4% SDS
0.2 M DTT
0.02% bromophenol blue
Extraction buffer
50 mM Tris-HCl, pH 7.5
25 mM NaCl
2 mM MgCl2
1 mM EDTA
5% glycerol
2 mM PMSF (add just before use)
Washing buffer
50 mM Tris-HCl, pH 7.5
800 mM NaCl
2 mM MgCl2
1 mM EDTA
5% glycerol
2 mM PMSF (add just before use)
Reaction Buffer
50 mM Tris-HCl, pH 7.5
25 mM NaCl
2 mM MgCl2
1 mM EDTA
2 mM dithiothreitol (DTT)
5% glycerol
50 μM Suc-LLVY-AMC substrate
Acknowledgments
The work was supported by grants from the NIH (grant GM127742) to X.Z. The protocol presented here was developed from several previous publications (Yang et al., 2004; Zhang et al., 2006; Book et al., 2010; Hsieh et al., 2015; Han et al., 2019) and optimized in our recent work (Li et al., 2020).
Competing interests
The authors declare no competing interests.
References
- Ben-Nissan, G. and Sharon, M. (2014). Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 4(3): 862-884.
- Book, A. J., Gladman, N. P., Lee, S. S., Scalf, M., Smith, L. M. and Vierstra, R. D. (2010). Affinity purification of the Arabidopsis 26S proteasome reveals a diverse array of plant proteolytic complexes. J Biol Chem 285: 25554-25569.
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Han, J. J., Yang, X., Wang, Q., Tang, L., Yu, F., Huang, X., et al. (2019). The β5 subunit is essential for intact 26S proteasome assembly to specifically promote plant autotrophic growth under salt stress. New Phytol 221(3): 1359-1368.
- Hsieh, L. S., Su, W. M., Han, G. S. and Carman, G. M. (2015). Phosphorylation regulates the ubiquitin-independent degradation of yeast Pah1 phosphatidate phosphatase by the 20S proteasome. J Biol Chem 290(18): 11467-11478.
- Leggett, D. S., Glickman, M. H. and Finley, D. (2005). Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. In: Ubiquitin-Proteasome Protocols. Springer pp: 57-70.
- Leggett, D. S., Hanna, J., Borodovsky, A., Crosas, B., Schmidt, M., Baker, R. T., et al. (2002). Multiple associated proteins regulate proteasome structure and function. Molecular cell 10(3): 495-507.
- Li, Y., Sun, D., Ma, Z., Yamaguchi, K., Wang, L., Zhong, S., et al. (2020). Degradation of SERRATE via ubiquitin-independent 20S proteasome to survey RNA metabolism. Nature Plants 6: 970-982.
- Reidlinger, J., Pike, A. M., Savory, P. J., Murray, R. Z. and Rivett, A. J. (1997). Catalytic properties of 26 S and 20 S proteasomes and radiolabeling of MB1, LMP7, and C7 subunits associated with trypsin-like and chymotrypsin-like activities. J Biol Chem 272(40): 24899-24905.
- Verma, R., Chen, S., Feldman, R., Schieltz, D., Yates, J., Dohmen, J., et al. (2000). Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 11(10): 3425-3439.
- Yang, P., Fu, H., Walker, J., Papa, C. M., Smalle, J., Ju, Y. M., et al. (2004). Purification of the Arabidopsis 26 S proteasome biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem 279(8): 6401-6413.
- Zhang, X. R., Henriques, R., Lin, S. S., Niu, Q. W. and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1(2): 641-646.
Article Information
Copyright
© 2021 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Li, Y., Sun, D., Yan, X., Wang, Z. and Zhang, X. (2021). In vitro Reconstitution Assays of Arabidopsis 20S Proteasome. Bio-protocol 11(7): e3967. DOI: 10.21769/BioProtoc.3967.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Activity
Molecular Biology > Protein > Targeted degradation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








