- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Live Cell Imaging of Male Meiosis in Arabidopsis by a Landmark-based System
(*contributed equally to this work) Published: Vol 10, Iss 9, May 5, 2020 DOI: 10.21769/BioProtoc.3611 Views: 6904
Reviewed by: Zinan ZhouLusheng FanAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2312 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1689 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 749 Views
Abstract
Live cell imaging has tremendously promoted our understanding of cellular and subcellular processes such as cell division. Here, we present a step-by-step protocol for a robust and easy-to-use live cell imaging approach to study male meiosis in the plant Arabidopsis thaliana as recently established. Our method relies on the concomitant analysis of two reporter genes that highlight chromosome configurations and microtubule dynamics. In combination, these reporter genes allowed the discrimination of five cellular parameters: cell shape, microtubule array, nucleus position, nucleolus position, and chromatin condensation. These parameters can adopt different states, e.g., the nucleus position can be central or lateral. Analyzing how tightly these states are associated gives rise to landmark stages that in turn allow a quantitative and qualitative dissection of meiotic progression. We envision that such an approach can also provide valuable criteria for the analysis of cell differentiation processes outside of meiosis.
Keywords: Cell divisionBackground
Meiosis is a special cell division cycle that serves two major purposes. First, the DNA content of the meiotic mother cell is reduced by half that leads, in the case of a diploid organism, to haploid meiotic products. This reduction is necessary for sexually reproducing organisms so that, after the fusion of two gametes during fertilization, the original genome size is restored. Second, meiosis promotes genetic diversity through the exchange of DNA segments between the parental chromosomes (homologous chromosomes or shortly homologs), named meiotic recombination, and by the generation of new, yet complete chromosome sets in which randomly either the homolog of the mother or the father is present for each chromosome (in case of a diploid organism). Thus, understanding meiosis is interesting for different fields of research ranging from cell biology and reproductive biology via genetics to evolutionary biology (Wijnker and Schnittger, 2013; Mercier et al., 2015; Melamed-Bessudo et al., 2016; Lambing and Heckmann, 2018; Pelé et al., 2018; Wang and Copenhaver, 2018).
Plants are a powerful model system to study the various aspects of meiosis. In addition, research in plant meiosis is also promoted by an applied interest since a low level of recombination often restricts breeding programs.
Research on plant meiosis has largely relied on the analysis of fixed cells, e.g., for chromosome spreads and immunolocalization studies, both for Arabidopsis (Armstrong et al., 2009; Martinez-Garcia and Pradillo, 2017; Parra-Nunez et al., 2020; Sims et al., 2020) as well as for other plant species (Chelysheva et al., 2013; Sepsi et al., 2018; Darrier et al., 2020; Stack et al., 2020). These techniques have been and continue to be important tools as they offer a great spatial resolution to address the structure of meiotic chromosomes and the localization of meiotic regulators for instance demonstrated by an immuno-cytological analysis of the 3D configuration of meiotic chromosomes (Hurel et al., 2018). However, these studies provide little to no information about the underlying dynamics of meiosis. In addition, temporal aspects of meiosis have to be indirectly deduced by the frequency of observed stages, a procedure that is inherently error-prone and can easily misguide the researcher e.g., when two or more populations of meiocytes exist in the same sample undergoing an altered course of meiosis that can be mistaken as one population with cells at different stages (Prusicki et al., 2019; Sofroni et al., unpublished). Moreover, short-lived phases, for instance nuclear envelope breakdown, are difficult to catch, and there has been for instance a long discussion in the field whether the nuclear envelop is reformed in Arabidopsis after the first meiotic division.
Live cell imaging of meiosis can complement the analyses of fixed material and build together with these techniques a powerful approach to reach molecular mechanistic insights into this important cell division program. Live cell-imaging of plant meiosis has been previously initiated in maize (Yu et al., 1997; Sheehan and Pawlowski, 2009; Nannas et al., 2016). However, only short phases of meiosis in maize could be recorded with a genetic reporter for microtubules and a chemical stain that highlights DNA. In addition, a recent protocol for live cell imaging of meiosis by light-sheet microscopy has been published (Valuchova et al., 2020). While this set-up is very powerful to follow meiosis in entire flower buds, it still does not reach the subcellular resolution as obtained by confocal laser scanning microscopy (CLSM).
Here, we describe in detail a method to follow meiosis in Arabidopsis anthers based on a recently established procedure by CLSM (Prusicki et al., 2019). Importantly, this method allows keeping the samples alive up to several days allowing the analysis of meiosis in its entirety. Furthermore, the use of Arabidopsis allowed the generation of plants containing fluorescent reporters for different meiotic regulators, foremost the meiosis-specific kleisin subunit of the cohesin complex called RECOMBINATION 8 (REC8), ASYNAPTIC 1 (ASY1) and ASY3, two components of the chromosome axis, and ZYP1, a component of the central region of the synaptonemal complex (Prusicki et al., 2019; Yang et al., 2019 and 2020). This set up has recently been used to study the control of cohesin in mutants in which REC8 is prematurely cleaved (Cromer et al., 2019).
A major challenge is the quantitative analysis of the obtained movies. As typical for biological processes, meiosis is a continuous and gradual succession of events. To dissect these movies, we focused on five cellular parameters as visualized by the KINGBIRD reporter system (REC8 labeled with GFP in combination with microtubules highlighted in red; Prusicki et al., 2019). These five cellular parameters are: cell shape, microtubule array, nucleus position, nucleolus position and chromatin state. Each of these parameters can adapt different states, see Table 1 (Table 1 Excel file), this publication and Figure 3 in Prusicki et al., 2019.
Looking then at the association of these different parameter states revealed that they are not randomly associated but often tightly linked. This gives rise to a biological landmark system where one landmark is a prominent cellular configuration with distinct parameter states. In turn, this landmark system can be used as a map to qualitatively (appearance of the same or new landmarks) and quantitatively (duration of these landmarks) dissect meiotic progression in mutants or different environmental conditions. The use of other meiotic reporters can then be used to refine and/or complement this landmark system. The principle of this analysis can be easily translated to other cellular differentiation processes, of course including other cellular parameters, which need to be identified.
Materials and Reagents
- Black marker
- Squared Petri dishes 100 x 100 x 20 mm (Sardstedt, catalog number: 82.9923.422 )
- Round Petri dishes 35 x 10 mm (Sarstedt, catalog number: 82.1135.500 )
- Round Petri dishes 60 x 15 mm (Sarstedt, catalog number: 82.1194.500 )
- Safeseal tube 1.5 ml (Sarstedt, catalog number: 72.706 )
- Parafilm M (Neolab, Bemis, 3-1011, PM-996)
- Sterilizing filter Millex-GV 0.22 μm (Merck, catalog number: SLGV033RS )
- Needle 30 G ½” 0.3 x 13 mm (BD Microlance, model: 304000 )
- Seeds of Arabidopsis thaliana plants containing meiotic fluorescent reporter constructs as a hallmark of meiosis such as the KINGBIRD line presented in Prusicki et al., 2019 as well as reporters for the meiotic chromosome axis and the synaptonemal complex (Yang et al., 2019 and 2020)
- NaClO in H2O, solution 13% (Applichem GmbH, ITW Reagents, catalog number: 213322.0715 )
- HCl 37% (VWR Chemicals, catalog number: 20255.29 0)
- Agarose (Sigma, catalog number: A9539 )
- Agar, powdered food grade (Applichem GmbH, ITW Reagents, catalog number: A00917,5000 )
- MS Basal Salt Mixture (Duchefa Biochemie, catalog number: M0221.0050 )
- Myo-Inositol (Duchefa Biochemie, catalog number: I0609.0100 )
- Nicotinic acid (Duchefa, catalog number: N0611 )
- Pyridoxin hydrochloride (Duchefa, catalog number: P0612 )
- Thiamine hydrochloride (Duchefa, catalog number: T0614 )
- Glycine (Sigma, catalog number: G-7126 )
- Sterile dH2O
- Acetocarmine
- Isopropanol
- Sucrose
- KOH
- Murashike and Skoog medium (MS medium) for plant germination (see Recipes)
- Arabidopsis Apex Culture Medium (ACM) for imaging (see Recipes)
Equipment
- Pipettes 10/100/1000 (e.g., Eppendorf Research plus pipette)
- Growth chambers MobyLux GroBanks (CLF Plant Climatics, model: BrightBoy )
- Vacuum chamber
- Glass bicker
- Clean bench
- Fine tweezers, thickness: 0.02/0.01 mm (Dumont, Dumoxel, catalog numbers: 0203-4-PO and 0203-5-PO )
- Dissection microscopes (e.g., Zeiss, models: Stemi2000 and Stemi508)
- Laser Scanning Microscope (e.g., Zeiss, model: LSM880 ), equipped with a water dipping objective (e.g., Zeiss, model: W Plan-Apochromat 40x/1.0 DIC objective )
Software
- ZEN 2.3 SP1 FP1 black (Zeiss) (or the related imaging software of your microscope system)
- MetaMorph Version 7.8.0.0 (Molecular devices)
- Fiji version 1.52b (ImageJ, https://imagej.net/Fiji) (Schindelin et al., 2012)
- Excel (Microsoft Office)
- Python programming language (Version 3.6, Python Software Foundation)
Procedure
- Preparation of plant material
- Surface-sterilize seeds of Arabidopsis thaliana, carrying a fluorescent reporter expressed at meiotic stages (an example is available in Prusicki et al., 2019). One efficient method to surface-sterilize seeds is by chloride gas: pour the seeds in 1.5 ml tubes, and mark the tubes with a black marker (other colors will faint due to the sterilization process). Leave the lid of the tubes open, and place the tubes in a racket inside a vacuum chamber. In the same chamber place a glass bicker containing 25 ml of a 7.5% bleach solution (eluted with tap water). Add to the bicker 1 ml of 37% HCl, quickly close the chamber and apply vacuum. The whole procedure should be carried out under a chemical hood. Be careful not to breathe the chloride gas! Leave the seeds in the chamber for 3-4 h. After the treatment, the seeds are ready to be sawn. If instead you are planning to store the sterilized seeds in the tube for some time, let the tubes open under a clean bench for a minimum of 30 min to ensure the gas has completely evaporated.
- Saw the seeds on Murashike and Skoog (½ MS, Recipe 1) squared Petri dishes enriched with selecting agent according with the resistance of the inserted construct. e.g., add 25 mg/L Hygromycin B to select for PROREC8:REC8:GFP as done in Prusicki et al. (2019). Alternatively, the selection can be performed by genotyping or based on fluorescence detection, if the protein of interest is expressed in seedlings.
- Store the plates for stratification in the dark for 3 days at 4 °C and then transfer them to long-day conditions (16 h day/8 h night regime at 22 °C/18 °C) for germination.
- 10 days after germination transfer the seedlings on soil and grow them at long-day conditions.
- Plants will be ready to image in 3 to 4 weeks, depending on growth conditions and genotypes.
- Preparation of imaging samples
- Prepare Apex Culture Medium (ACM) with agarose to 0.8%, 1% and 2% concentrations and 1,000x vitamin stock as indicated in the session (Recipe 2).
- Sterilize the ACM by autoclaving.
- When the autoclaved (or reheated) ACM with 0.8% agarose is hand warm, add the vitamin mix to a 1x working concentration (e.g., to 50 ml of ACM add 50 μl of a 1,000x vitamin stock solution), and pour it into small Petri dishes (35 x 15 mm), seal with parafilm and store at 4 °C.
- Pour the ACM with 1% agarose in medium-sized Petri dishes (60 x 15 mm), seal with parafilm and store at 4 °C.
- Aliquot the ACM with 2% agarose in sterile 1.5 ml Eppendorf tubes and store them at 4 °C.
- Sample mounting
- Cut an inflorescence and anchor it on a Petri dish with ACM with 1% agarose (Video 1).Video 1. Removal of large flower buds and selection of the flower bud for image acquisition, Steps C1 and C2
- Remove all the open and the elongated flowers at the base of the pedicel but the flower bud with a roundish shape and the size between 0.4-0.6 mm containing early meiocytes (Video 1). The characteristics of flower buds with meiocytes in an early stage can vary depending on the growth conditions and the genotype, e.g., mutants in which meiosis is delayed usually takes places in flower buds of larger sizes. It is therefore recommended to analyze several flower buds with acetocarmine or similar staining to estimate in which flower buds meiotic stages can be found. In general, flower buds at developmental stage 9 (Smyth et al., 1990) contain male meiocytes undergoing meiosis.
- Remove the upper sepal of the selected flower bud, being careful to not touch the inner organs of the flower, in particular the anthers (Videos 2 and 3).Video 2. Removal of sepal to give access to two anthers in the selected flower bud, Steps C3 and C4Video 3. Close up of a flower bud ready to image, after Steps C3 and C4
- Remove the remaining flower buds (Videos 2 and 3). The next smallest/youngest flower buds can be left attached as a backup flower in the case the main flower bud gets damaged during the removal of the sepal.
- Lift the inflorescence from the preparation medium and cleanly cut the stem to a length of circa (ca.) 0.5 cm (with forceps or better using a needle) (Video 4). This will facilitate the uptake of nutrients from the medium and will keep the sample in good condition for a long time.Video 4. Positioning of the selected and processed flower bud for imaging, steps C5 and C6
- Transfer the sample onto the small Petri dish with ACM with 0.8% agarose, anchor it in the middle of the plate and cover the flower bud with a drop of ACM with 2% agarose (Video 4 and Figures 1A and 1B).
- It is possible to follow more than one flower bud in one microscopy session. In this case, prepare a new sample as previously explained (starting from Procedure C1 of the Bio-protocol), and locate all flower buds as close as possible in the center of the Petri dish (Video 4).
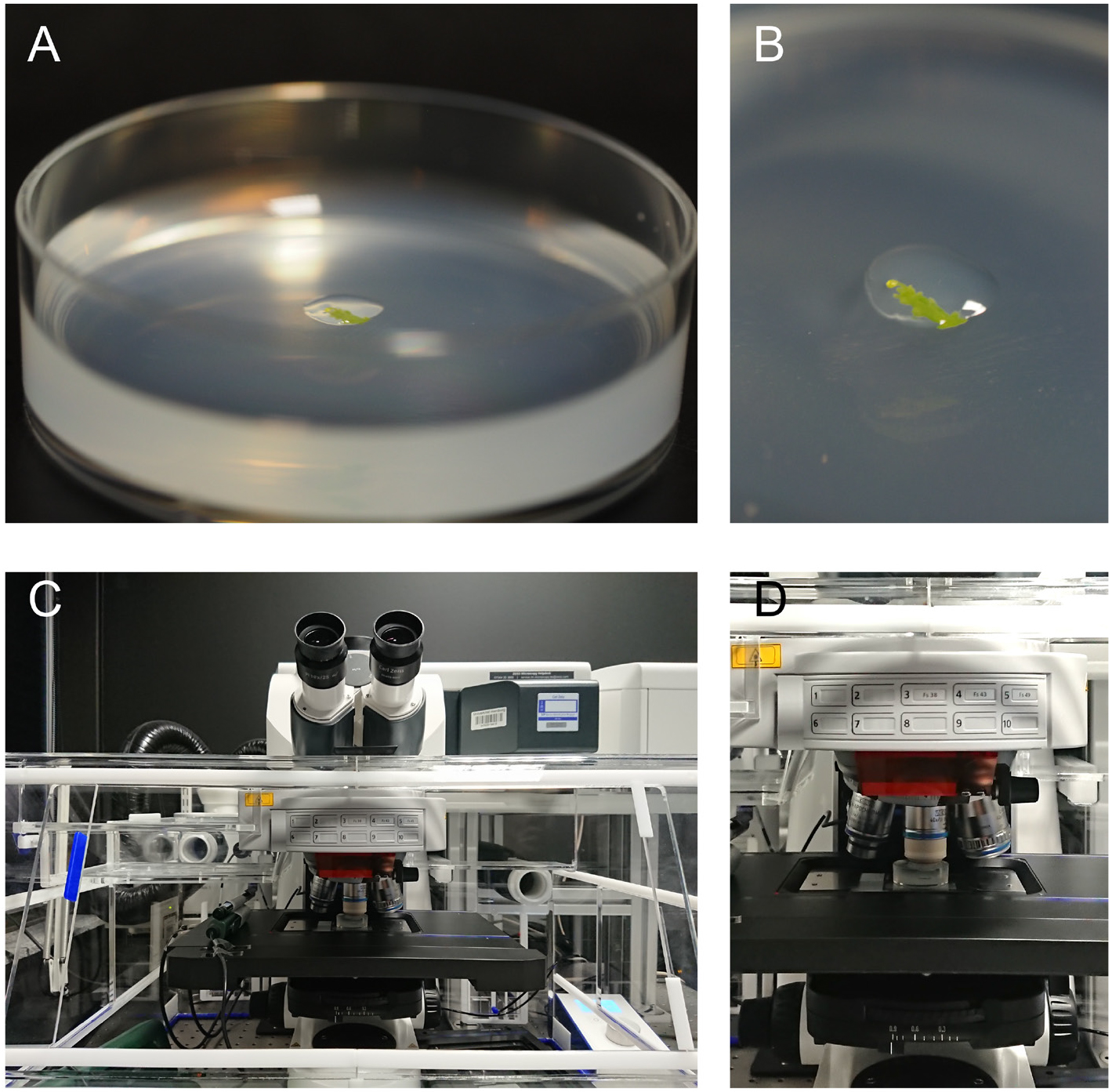
Figure 1. Sample mounting and positioning at the microscope. A. The sample is placed in the middle of a Petri dish and covered by a drop of 2% agarose. B. Close up of the sample from panel A. C Overview and D zoom onto the microscope stage with the sample positioned under the water immersion objective.
- Cut an inflorescence and anchor it on a Petri dish with ACM with 1% agarose (Video 1).
- Image acquisition
- It is important to have stable acquisition conditions, e.g., room temperature, humidity, etc. Therefore, it is advisable to turn on the confocal microscope in advance and position the sample in the room with a few ml of water covering the mounted samples.
- After approximately one hour you can start the image acquisition.
- Position the sample on the microscope stage, submerge the water-dipping objective and fill up the Petri dish to the top with autoclaved water (Figures 1C and 1D). It is advisable to thoroughly clean the objective with isopropanol before the acquisition to reduce the risk of bacteria growth during image acquisition.
- Using the visual function (or related software tools), identify the position of each flower bud in the sample and save it using the multi-position function on ZEN black software.
- The set-up for image acquisition needs to be adapted to the purpose of the experiment and to the characteristics of the used fluorophores. To follow a complete meiotic division in a time-lapse experiment, and to identify the meiotic landmarks using the KINGBIRD line as presented in Prusicki et al. (2019), the following set-up can be used:
- Use the Argon laser at λ 488 nm to excite GFP and the DPSS 561-10 laser (λ 561) to excite TagRFP.
- Use two tracks in sequential line mode for signal detection, and the Beam splitter MBS 488/561. In the first track, use the GaAsP detector to record the GFP signal filtered for λ between 498 and 550 nm. In the second track, use the GaAsP detector to observe the TagRFP signal filtered for λ between 578 and 649 nm. A third channel to detect the autofluorescence of chloroplast can be added to the second track and filtered for λ between 680 and 750 nm. Additionally, the Transmitted Light detection can be added to one of the two tracks.
- Set the Pinhole at 1 Airy Unity for the TagRFP detection, use bidirectional scan function and set the pixel dwell to a value around 2 μs.
- Parameters for image acquisition such as laser intensity, gain and offset have to be adapted to the individual imaging conditions. Among others, they depend on the laser status, as well as on the reporter line used, and thus the level of protein expression. In general, a compromise between sample viability and high-resolution imaging has to be found to obtain the best image quality while maintaining the sample in good conditions. In our case, we set the intensity of the Argon laser between 1% and 4.5%, while the DPSS 561-10 laser was set between 0.3% and 1%. The detector gain to collect GFP and TaqRFP signals was set between 700 and 850, and was set between 650 and 750 for the detection of chloroplast autofluorescence. In all the cases, the offset parameter was 0.
- Perform averaging on 2 lines in case of the KINGBIRD line; for other reporters, different setting may be needed.
- Acquire time lapse of multiple positions with 10 min interval time. At each position, perform a Z-stack of 6 to 7 planes with a step size to a maximum of 50 μm per time point. To follow the entire meiotic division, images should be acquired for ca. 30 h (Prusicki et al., 2019).
- The use of the Autofocus function at each position and each time point is suggested, especially for long acquisition times.
- In case the sample is still going through meiosis but the data acquisition needs to be shortly interrupted (e.g., in case of readjustment of the focal plane or in case of reaching the last acquisition cycle) it is advisable to fit the time of interruption to match the interval time, so to start the new time-lapse acquisition at the end of the 10 min interval.
- Image processing
- Save each position of the acquired time-lapse as a separate file, for instance in the case of the ZEN black software: processing → copy→ subset.
- Open the image in Fiji and save it in a separate folder as image sequence (save as → image sequence → format TIFF → digits must be enough to cover all time points e.g., 110 time points need 3 digits).
- Open the image sequence using MetaMorph (App → review multiple image sequence file → open sequence → follow the instructions), and select for each time point the z-stack layer, which shows the cells of interest. Finally, load the selected z-stacks for each wavelength and save them as tiff. files. This step helps to correct for the drift of the sample in the z-direction, and together with the autofocus function activated during image acquisition (see Procedure D6) allows following the same cells for the complete duration of the time lapse.
- Open the new files in Fiji, combine the different channels, adjust their brightness and contrast and generate an RGB version of the file.
- If an x-y drift is detectable, it can be corrected by using the plug-in Stackreg from Fiji (Plugins → Stackreg → Rigid body) (Thévenaz et al., 1998).
- An example of a time-lapse series after image processing is given in Video 5.Video 5. Example of the outcome of a time lapse experiment after image processing as explained in Steps E1-E6
Data analysis
Landmark identification using the KINGBIRD line
- Data set annotation
- Give an ID number to each visible cell within one anther. The ID number could be impressed on each frame of the RGB.tiff file using the Text tool of Fiji (Figure 2).
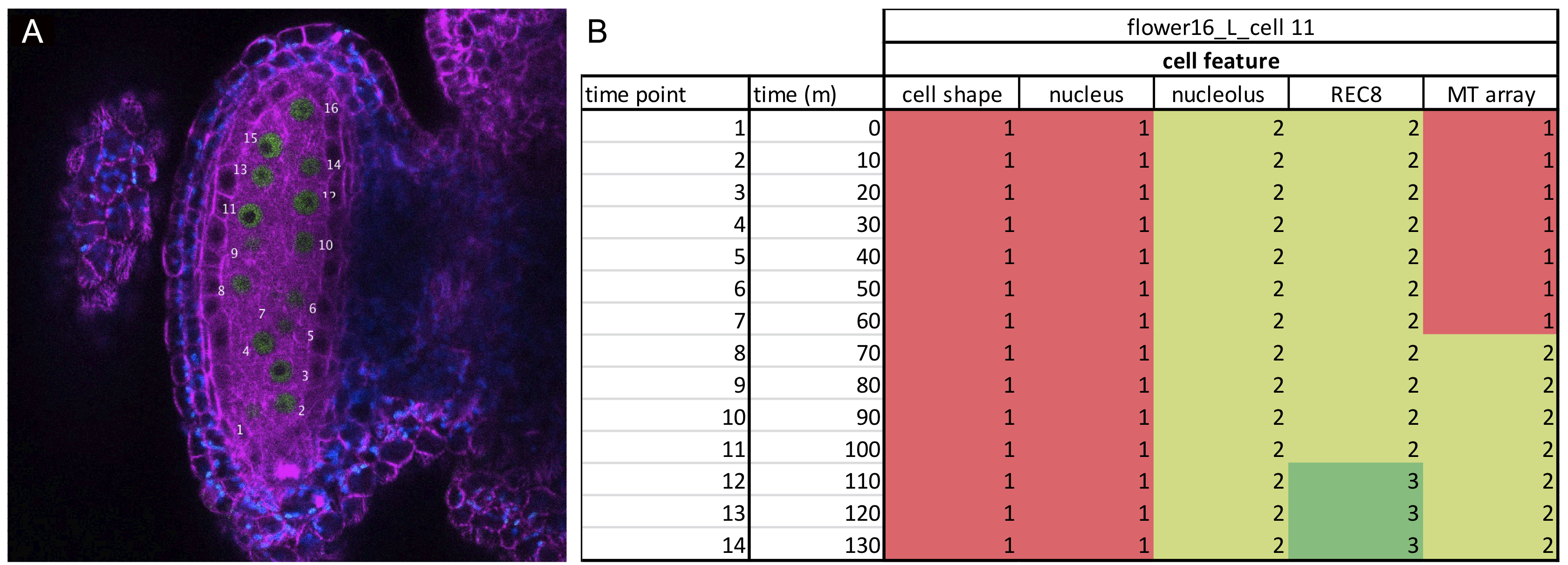
Figure 2. Data analysis. A. One frame of an acquired time-lapse movie depicting an anther of an Arabidopsis plant carrying the KINGBIRD reporter (microtubules are in magenta, REC8-chromatin is in green and chloroplasts are blue). Each meiocyte is given a number, which will be its ID on the spreadsheet in B. B. Example of a spreadsheet used for analysis as described in the paragraph Data-analysis A2. The numbering of the stage refers to the numbers in Table 1. Each stage is color-coded to facilitate the data organization. - Prepare a table using Microsoft Excel that includes 7 columns: frame number, time in min, cell shape, microtubule array, nucleus position, nucleolus position and chromatin state (Figure 2). The number of rows will be equal to the number of frames acquired for a single cell. The first imaged frame will be counted as Time 0 (Figure 2).
- Fill in each column with a number that corresponds to the state of the analyzed cellular parameter as annotated in Table 1 and Figure 2 (for a further reference see Prusicki et al., 2019). For instance, if at time 0 the cell presents a round shape, the number 4 must be filled in in the third column. Proceed with the annotation of each cellular parameter at each time point. If the cell is not visible at one time point, or the parameter state cannot be recognized, fill in the table an n for non-visible.
- Annotate all the visible cells of one another on different Excel sheets (one sheet per cell). This file constitutes the data set that will be analyzed as explained in the following session.
Table 1. Cell features of meiocytes as obtained with the KINGBIRD reporter system. The table provides microscopy images that illustrate each described cell feature (adapted from Figures 3 and 4 in Prusicki et al., 2019. Creative Commons Attribution License). The original table file could be accessed here (Table 1 Excel file).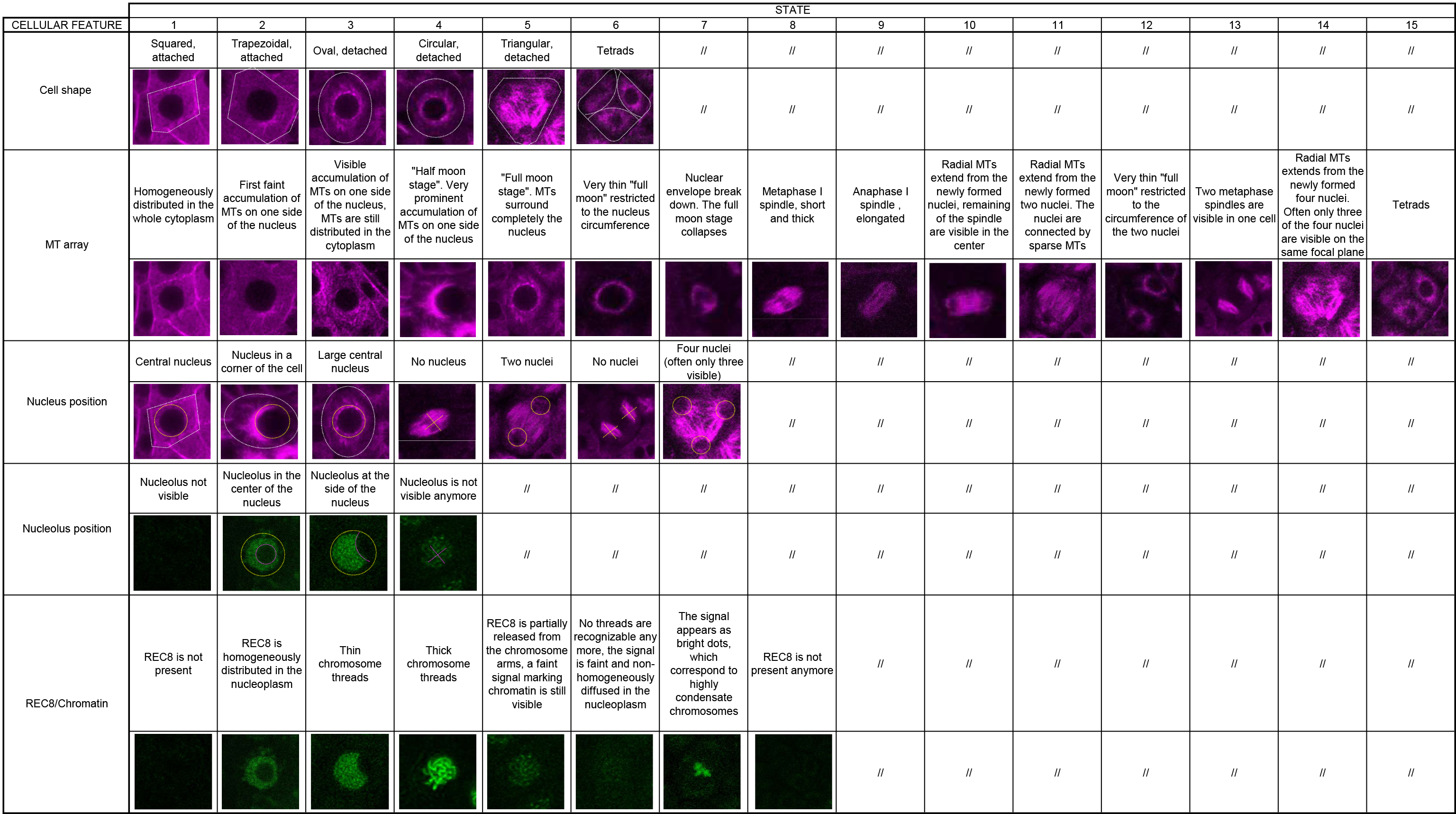
- Give an ID number to each visible cell within one anther. The ID number could be impressed on each frame of the RGB.tiff file using the Text tool of Fiji (Figure 2).
- Data set analysis
A Python script to perform analysis of the annotated data set created in part A. is available at https://gitlab.com/wurssb/arabidopsis-thaliana---landmark-analysis. This script contains the procedure to extract landmarks from the data and takes the following steps:- Prepare the data for analysis by filtering out invalid or duplicate entries. Each entry, consisting of the observed state for each cellular parameter should have an identifier for the sample and the specific cell within the sample.
- If the data is not sampled at regular intervals, resample to a common interval. However, take care when resampling, as going from short to large intervals might remove short-lived states from the data set while the reverse could introduce (large) errors from interpolating the states.
- A cellular state is defined by the ensemble of all parameter states. For example, cell 11 from Figure 2 at time point 10 has the cellular state: 1-1-2-2-2. Cellular states diverging for up to two different parameter states were defined as neighboring states (or neighbors). Hence, the cellular states 1-1-2-2-1, 1-1-2-3-2 and 1-1-2-3-3 but not 1-1-3-3-3 are neighboring states to 1-1-2-2-2.
- Partially observed states can be either discarded, or interpolated from neighboring states. Again, take care when interpolating, for example, by setting a maximum time interval that can be safely interpolated depending on the observed processes.
- Calculate how much time is spent in each cellular state.
- For each observed cellular state, list the neighboring states.
- For each cellular state, calculate the neighboring score using the time spent in the specific state (tstate) and the time spent its neighbors (tneighbors):
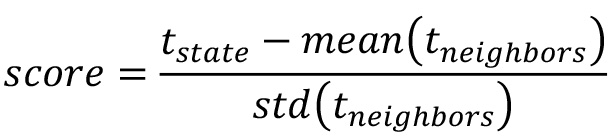
- Sort the cellular states by their score. Landmarks can now be assigned with a suitable cut-off value (an example in Prusicki et al., 2019). An example of the final result of the neighboring analysis is given in Table 2 (Table 2 Excel file), where all the cellular states observed are listed together with their counts (number of cases they have been observed), their percentage of appearance in relation to the other cellular states and finally their neighboring score. The source of the data listed in Table 2 are the results published in Prusicki et al., 2019.
- To assess the robustness of the analysis, bootstrap the analysis by resampling the data with replacement and re-running the whole analysis multiple times.
Bootstrapping can be done on the level of single observations, or by resampling groups of observations of specific cells or anthers to uncover different sources of possible bias in the analysis.
Table 2. Example of an outcome of the data analysis. The original table file could be accessed here (Table 2 Excel file).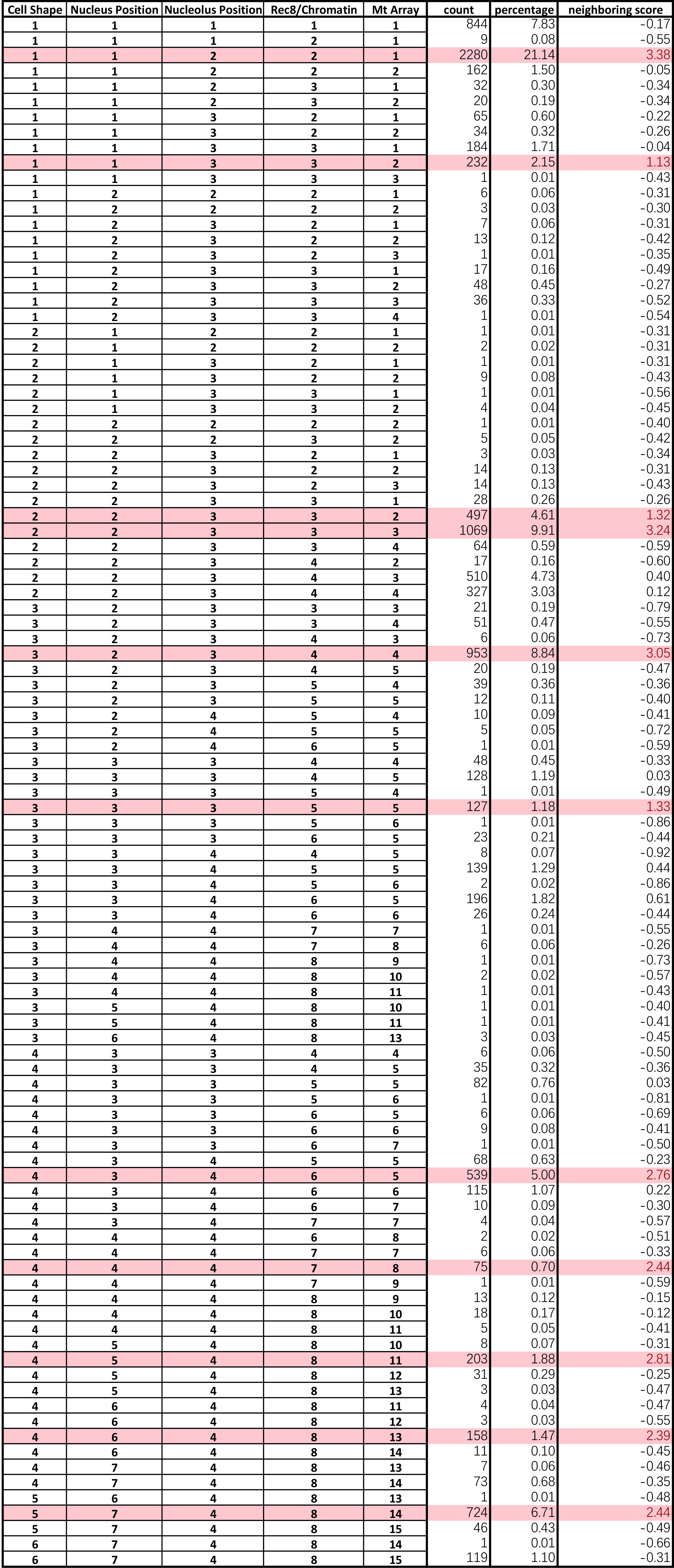
Recipes
- Murashike and Skoog medium (MS medium) for plant germination
Ingredient Concentration For 1 L MS basal medium 0.5x 2.2 g/L Sucrose 1% 10 g/L Agar 1% 10 g dH2O --- up to 1 L of volume KOH --- --- - Dissolve MS basal and sucrose in 800 ml of dH2O
- Adjust the pH to 5.8 using KOH and bring to the final volume of 1 L
- Add the agar to the solution
- Sterilize by autoclaving and store at room temperature until usage
- Arabidopsis Apex Culture Medium (ACM) for imaging
Ingredient Concentration Quantity MS-Medium base MS basal medium 0.5x 2.2 g/L Sucrose 1% 10 g/L Agarose 0.8%-1%-2% 8 g-10 g-20 g dH2O --- up to 1 L of volume KOH --- --- Vitamins (1,000x) Myo-Inositol 10% 5 g/50 ml Nicotinic acid 0.1% 0.05 g/50 ml Pyridoxin hydrochloride 0.1% 0.05 g/50 ml Thiamine hydrochloride 1% 0.5 g/50 ml Glycine 0.2% 0.1 g/50 ml dH2O --- 50 ml - Prepare the MS base as described in the previous recipe, being careful to add agarose to the three different concentrations (0.8%, 1% and 2%) instead of agar and store it at room temperature
- Dissolve the vitamins in dH2O to obtain the 1,000x stock solution
- Sterilize the vitamin stock using a 0.22 μm filter, aliquot them and store at -20 °C
Acknowledgments
This work was supported by the European Union Marie-Curie “COMREC” network FP7 ITN-606956 to M.A.P. and A.S. In addition, core funding of the University of Hamburg to A.S. is gratefully acknowledged. The here-presented protocol was presented in Prusicki et al. (2019) and further described in Prusicki et al. (2020).
Competing interests
Arp Schnittger on behalf of all authors confirms that none of the authors has competing financial interests in this work.
References
- Armstrong, S. J., Sanchez-Moran, E. and Franklin, F. C. (2009). Cytological analysis of Arabidopsis thaliana meiotic chromosomes. Methods Mol Biol 558: 131-145.
- Chelysheva, L. A., Grandont, L. and Grelon, M. (2013). Immunolocalization of meiotic proteins in Brassicaceae: method 1. Methods Mol Biol 990: 93-101.
- Cromer, L., Jolivet, S., Singh, D. K., Berthier, F., De Winne, N., De Jaeger, G., Komaki, S., Prusicki, M. A., Schnittger, A., Guerois, R. and Mercier, R. (2019). Patronus is the elusive plant securin, preventing chromosome separation by antagonizing separase. Proc Natl Acad Sci U S A 116(32): 16018-16027.
- Darrier, B., Arrieta, M., Mittmann, S. U., Sourdille, P., Ramsay, L., Waugh, R. and Colas, I. (2020). Following the formation of synaptonemal complex formation in wheat and barley by high-resolution microscopy. Methods Mol Biol 2061: 207-215.
- Hurel, A., Phillips, D., Vrielynck, N., Mézard, C., Grelon, M. and Christophorou, N. (2018). A cytological approach to studying meiotic recombination and chromosome dynamics in Arabidopsis thaliana male meiocytes in three dimensions. Plant J 95(2): 385-396.
- Lambing, C. and Heckmann, S. (2018). Tackling plant meiosis: from model research to crop improvement. Front Plant Sci 9: 829.
- Martinez-Garcia, M. and Pradillo, M. (2017). Functional Analysis of Arabidopsis ARGONAUTEs in Meiosis and DNA Repair. Methods Mol Biol 1640: 145-158.
- Melamed-Bessudo, C., Shilo, S. and Levy, A. A. (2016). Meiotic recombination and genome evolution in plants. Curr Opin Plant Biol 30: 82-87.
- Mercier, R., Mezard, C., Jenczewski, E., Macaisne, N. and Grelon, M. (2015). The molecular biology of meiosis in plants. Annu Rev Plant Biol 66: 297-327.
- Nannas, N. J., Higgins, D. M. and Dawe, R. K. (2016). Anaphase asymmetry and dynamic repositioning of the division plane during maize meiosis. J Cell Sci 129(21): 4014-4024.
- Parra-Nunez, P., Pradillo, M. and Santos, J. L. (2020). How to perform an accurate analysis of metaphase I chromosome configurations in autopolyploids of Arabidopsis thaliana. Methods Mol Biol 2061: 25-36.
- Pelé, A., Rousseau-Gueutin, M. and Chevre, A. M. (2018). Speciation success of polyploid plants closely relates to the regulation of meiotic recombination. Front Plant Sci 9: 907.
- Prusicki, M. A., Keizer, E. M., van Rosmalen, R. P., Komaki, S., Seifert, F., Muller, K., Wijnker, E., Fleck, C. and Schnittger, A. (2019). Live cell imaging of meiosis in Arabidopsis thaliana. Elife 8: 42834 .
- Prusicki, M. A., Hamamura, Y. and Schnittger, A. (2020). A practical guide to live-cell imaging of meiosis in Arabidopsis. Methods Mol Biol 2061: 3-12.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J. Y., White, D. J., Hartenstein, V., Eliceiri, K., Tomancak, P. and Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7): 676-682.
- Sepsi, A., Fabian, A., Jager, K., Heslop-Harrison, J. S. and Schwarzacher, T. (2018). Immunofish: simultaneous visualisation of proteins and DNA sequences gives insight into meiotic processes in nuclei of grasses. Front Plant Sci 9: 1193.
- Sheehan, M. J. and Pawlowski, W. P. (2009). Live imaging of rapid chromosome movements in meiotic prophase I in maize. Proc Natl Acad Sci U S A 106(49): 20989-20994.
- Sims, J., Chouaref, J. and Schlogelhofer, P. (2020). Whole-mount immuno-fish on Arabidopsis meiocytes (whomi-fish). Methods Mol Biol 2061: 59-66.
- Smyth, D. R., Bowman, J. L. and Meyerowitz, E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2(8): 755-767.
- Sofroni K., Takatsuka H., Yang, C., Dissmeyer N., Komaki, S., Hamamura, Y., Böttger, L., Umeda M. and Schnittger A. CDKD-dependent activation of CKDA;1 controls microtubule dynamics and cytokinesis during meiosis. (Unpublished).
- Stack, S. M., Shearer, L. A., Lohmiller, L. D. and Anderson, L. K. (2020). Preparing maize synaptonemal complex spreads and sequential immunofluorescence and fluorescence in situ hybridization. Methods Mol Biol 2061: 79-115.
- Thévenaz, P., Ruttimann, U. E. and Unser, M. (1998). A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7(1): 27-41.
- Valuchova, S., Mikulkova, P., Pecinkova, J., Klimova, J., Krumnikl, M., Bainar, P., Heckmann, S., Tomancak, P. and Riha, K. (2020). Imaging plant germline differentiation within Arabidopsis flowers by light sheet microscopy. Elife 9: e52546.
- Wang, Y. and Copenhaver, G. P. (2018). Meiotic recombination: mixing it up in plants. Annu Rev Plant Biol 69: 577-609.
- Wijnker, E. and Schnittger, A. (2013). Control of the meiotic cell division program in plants. Plant Reprod 26(3): 143-158.
- Yang, C., Hamamura, Y., Sofroni, K., Böwer, F., Stolze, S. C., Nakagami, H. and Schnittger, A. (2019). SWITCH 1/DYAD is a WINGS APART-LIKE antagonist that maintains sister chromatid cohesion in meiosis. Nat Commun 10(1): 1755.
- Yang, C., Sofroni, K., Wijnker, E., Hamamura, Y., Carstens, L., Harashima, H., Stolze, S. C., Vezon, D., Chelysheva, L., Orban-Nemeth, Z., Pochon, G., Nakagami, H., Schlogelhofer, P., Grelon, M. and Schnittger, A. (2010). The Arabidopsis Cdk1/Cdk2 homolog CDKA;1 controls chromosome axis assembly during plant meiosis. EMBO J: e101625.
- Yu, H. G., Hiatt, E. N., Chan, A., Sweeney, M. and Dawe, R. K. (1997). Neocentromere-mediated chromosome movement in maize. J Cell Biol 139(4): 831-840.
Article Information
Copyright
Prusicki et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Prusicki, M. A., Keizer, E. M., van Rosmalen, R. P., Fleck, C. and Schnittger, A. (2020). Live Cell Imaging of Male Meiosis in Arabidopsis by a Landmark-based System. Bio-protocol 10(9): e3611. DOI: 10.21769/BioProtoc.3611.
- Prusicki, M. A., Keizer, E. M., van Rosmalen, R. P., Komaki, S., Seifert, F., Muller, K., Wijnker, E., Fleck, C. and Schnittger, A. (2019). Live cell imaging of meiosis in Arabidopsis thaliana. Elife 8: 42834 .
Category
Developmental Biology > Morphogenesis > Cell structure
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










