- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assembly of a Custom-made Device to Study Spreading Patterns of Pseudomonas putida Biofilms
(*contributed equally to this work) Published: Vol 9, Iss 10, May 20, 2019 DOI: 10.21769/BioProtoc.3238 Views: 4885
Reviewed by: Alba BlesaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Inexpensive Imaging Platform to Record and Quantitate Bacterial Swarming
Weijie Chen [...] Jay X. Tang
Sep 20, 2021 3635 Views

Purification of the Bacterial Amyloid “Curli” from Salmonella enterica Serovar Typhimurium and Detection of Curli from Infected Host Tissues
Murugesan Sivaranjani [...] Aaron P. White
May 20, 2022 3233 Views

A Guideline for Assessment and Characterization of Bacterial Biofilm Formation in the Presence of Inhibitory Compounds
Bassam A. Elgamoudi and Victoria Korolik
Nov 5, 2023 3102 Views
Abstract
Biofilms are bacterial communities in the shape of exopolysaccharide matrix-encased aggregates attached onto interphases able to resist environmental aggressions. The development of bacteria in the shape of biofilms deeply affects the performance of many industrial processes which work with fluidic systems, where bacteria may settle and prosper. As a consequence industrial equipment experiments low performance issues and substantial maintenance costs.
The study of how bacteria of industrial interest such as Pseudomonas putida spread in these fluidic systems is highly dependent on the chosen experimental system to retrieve such data, thus using scaled prototypes becomes an essential step towards the design of a more efficient system to handle biofilms, either to control them or to prevent them. This protocol describes how to assemble, operate and maintain a device to grow and monitor the biofilm spreading pattern of this bacterium (as a function of the fluid hydrodynamics) in a custom-made chamber larger than those typically used in laboratory environments, and how to analyze the information gathered from it in a straightforward fashion. Description of the protocol was thought to be used as a working template not only for the presented case study but for any other potential experiment in different contexts and diverse scales following similar design principles.
Background
The characterization of bacterial biofilm spreading pattern dynamics in fluidic systems is a key issue when trying to gain control of the proliferation of these living organisms. Especially relevant is the case of biofilm presence in industrial environments, where an uncontrolled growth may lead to large economic loses. Typical studies on this area focus their efforts on creating laboratory-scale setups to investigate the effect of different variables to test. These parameters are controlled to monitor subsequent biofilm expansion for every tested experimental condition. However extrapolating these results to an industrial-like fluidic environment is not so straightforward, as equipment, materials and operation conditions may deeply affect biofilm behavior. Furthermore the designs performed for laboratory-scale devices are in many cases incompatible with typical industrial equipment and procedures. As a consequence, the number of studies in the literature using industrial-like devices is very limited.
In this protocol we describe a procedure for assembling, testing and operating an industrial-like device to study the biofilm spreading pattern of platform strain Pseudomonas putida mt-2. The design and operation procedures of the device were specifically chosen to have into account constrains and features typically found when using industrial materials and equipment, delivering a viable solution for given research purposes.
Materials and Reagents
- Laboratory consumables
- 0.4 μm pore size nitrile filter (Millipore, catalog number: SLHA033SS)
- 0.45 μm PES filter unit (VWR, catalog number: 514-0335)
- Microscope slide (VWR, catalog number: 16004-422)
- Microscope cover slip (VWR, catalog number: 48393-026)
- Aluminum foil
- Marker pen
- Tubing
- 1 x 25 m 5.5 mm inner diameter flexible polyurethane tubing (Legris, catalog number: 1025U0805)
- 1 x Silicone tubing, 25 m, 2 mm (id) x 4 mm (od), (VWR, catalog number: 228-0704)
- 1 x Silicone tubing, 4 m, 4 mm (id) x 6 mm (od), (VWR, catalog number: 228-0709)
- Valves and clamps
- 8 x F-F ¼’ BSPP ball valve (RS, RS Pro, catalog number: 733-5202)
- 1 x ISMATEC 2 stop coded tubes, id: 1.65 mm (blue-blue, catalog number: SC0018), Mat: Tygon R-3607 (Cole Parmer, catalog number: EW-96460-38)
- 2 x hosecock clamp (VWR, catalog number: 21716-102)
- Connectors and fittings
- 1 x 8 mm–8 mm–8 mm Tee Legris (RS, catalog number: 445-4495)
- 14 x Legris Pneumatic Straight Threaded-to-Tube Adapter, R 1/4" Male, Push In 8 mm (RS, catalog number: 182-4784)
- 3 x SMC Pneumatic Straight Threaded-to-Tube Adapter, Rc 1/8" Female, Push In 8 mm (RS, catalog number: 771-5002)
- 2 x SMC Pneumatic Elbow Threaded-to-Tube Adapter, Push In 8 mm 1/4" thread (RS, catalog number: 771-6030)
- 2 x Legris Pneumatic Elbow Threaded Adapter, R 1/4" Male x R 1/4" Male (RS, catalog number: 367-5792)
- 2 x Legris Brass 1/4" in. BSPT Male Plug Threaded Fitting (RS, catalog number: 231-5045)
- 1 x Legris Pneumatic Tee Threaded-to-Tube Adapter, 1/4". x 8 mm x 8 mm, 30 bar (RS, catalog number: 225-0617)
- 1 x Barbed fittings, Straight Connector, Natural PP, 1/8" ID; 10/pk (Cole-Parmer, catalog number: EW-51518-05)
- Barbed Tee Connector, Polypropylene, 1/8"; 25/pk (Cole-Parmer, catalog number: SI-50623-66)
- 1 x ADCF Male Luer to 1/8" L Barb Adapter, Polypropylene, 25/pk (Cole-Parmer, catalog number: SI-30800-24)
- 1 x ADCF Female Luer to 1/8" L Barb Adapter, Polypropylene, 25/pk (Cole-Parmer, catalog number: SI-30800-08)
- Accessories
- Electrical Tape, 19 mm x 20m (RS, catalog number: 511-4306)
- Socket extension lead (RS, catalog number: 122-1105)
- 1 x Clear Polycarbonate PC Sheet, 1.25 m x 610 mm x 6 mm (RS, catalog number: 681-665)
- 1 x Nitrile rubber O-Ring Cord, 2 mm diameter (RS, catalog number: 138-1600)
- 1 x White PTFE Tape 12 mm x 12 m x 0.075 mm (RS, catalog number: 512-238)
- 1 x Pan Head Bright Zinc Plated Steel Machine Screw bag, M3, 20 mm (RS, catalog number: 560-625)
- 1 x Stainless Steel, Hex Nut bag, M3 (RS, catalog number: 189-563)
- 1 x Stainless Steel Plain Washer, 0.5 mm Thickness, M3 (RS, catalog number: 189-620)
- 1 x Cyanoacrylate glue (RS, catalog number: 533-478)
- Chemicals
- Gentamycin (Sigma-Aldrich, catalog number: PHR1077-1G)
- Absolute Ethanol (Sigma-Aldrich, catalog number: 1009832500)
- NaClO (VWR, catalog number: 470302-586)
- Glycerol 85% (Sigma-Aldrich, catalog number: 1040942500)
- Na2HPO4·2H2O (Sigma-Aldrich, catalog number: 71643-250G)
- KH2PO4 (Sigma-Aldrich, catalog number: P5655-500G)
- NaCl (Sigma-Aldrich, catalog number: S3014-1KG)
- NH4Cl (Sigma-Aldrich, catalog number: A9434-500G)
- MgSO4 (Sigma-Aldrich, catalog number: M2643-500G)
- 10x M9 salts stock (see Recipes)
- 20% Glycerol (see Recipes)
- 1 M MgSO4 (see Recipes)
- M9 minimal medium supplemented with 0.2% (w/v) glycerol (see Recipes)
Equipment
- 2 x 2 L Pyrex® narrow-mouth Erlenmeyer flask (Sigma-Aldrich, catalog number: CLS49802L)
- 3 x 1 L Pyrex® narrow-mouth Erlenmeyer flask (Sigma-Aldrich, catalog number: CLS49801L)
- 1x DuranTM 4-Port Assembled HPLC Screw Caps GL-45 (Fisher Scientific, catalog number: 10740834)
- 1 x 1 L Laboratory bottles with screw cap, DURAN® GL-45 (VWR, catalog number: 215-1517)
- 1 x 5 L BRAND® PP beaker with spout, low form (Sigma-Aldrich, catalog number: BR87826-4EA)
- 1 x 16 mm length magnetic stirring bar (VWR, catalog number: 442-0366)
- 1 x 250 x 250 mm magnetic stirrer (VWR, catalog number: 444-0572)
- Orbital shaker
- 1 x Flowmeter (Aalborg P-150 mm, 1/4" FNTP fitting, catalog number: P11A3-BB0)
- 2 x 12VDC Pumps (EKWD, EK-DCP 2.2, catalog number: 3831109862506)
- 3 x 220V AC/12V DC adaptor (RS Online, catalog number: 737-8149)
- 1 x 19-37.8 L aquarium air pump (Hagen, A842 Aquaclear Air pump 10, catalog number: 15561108447)
- 1 x Ismatec IPC-N ISM936D IPC-N (ISM936D) Low-Speed Digital Peristaltic Pump with Click-'N-Go Cartridges; 8-Channel, 230V (Cole-Parmer, catalog number: EW-78001-16)
- 1 x Adjustable Spanner (RS, catalog number: 469-7018)
- 1 x EMCO Concept Mill 105 milling machine
- 1x Fluorescence microscope with the following features:
- Light source and light filter to allow visualization of GFP and Texas Red fluorochromes
- Monochrome digital camera with EMCCD chip (minimum requirement)
- Motorized tray for automatic XY displacement and automatic tile-scan acquisition
- Automatic Z autofocus
Software
- Scientific programming language (i.e., MATLAB) or general purpose programming language (i.e., Python, C+) able to work with scientific computing packages supporting basic statistic functions (i.e., mean, standard deviation), array/structure manipulation (array sorting, length) and element selection operators (unique elements)
- Image treatment library for selected program language (i.e., scikit-image for Python, Image processing toolbox for MATLAB)
- Image treatment software (ImageJ or equivalent) containing a proper library to open microscope files with native formats (i.e., *.lif), or microscope-related software able to perform regular format file storage
Procedure
The complete experiment could be divided into the following 6 sections (A to F).
- Flow cell/Biofilm chamber design and manufacturing
The design and manufacturing of the biofilm chamber involved the use of a milling machine to generate the geometry specified in Figure 1. The biofilm chamber geometry was designed with the aim of promoting a laminar flow through a rectangular square duct of variable Reynolds number, being this last parameter adjustable by setting the flow rate at the entrance. The central part of the chamber was divided in smaller ducts having the proper hydraulic diameter to create the hydrodynamic conditions to study within the channels (a laminar flow with Re~100-1,000). The ducts additionally served as a way of taking technical replica of different sections of the chamber placed at different positions. Two lateral chambers were also added at the input and output sides to hydrodynamically stabilize the fluid pattern before entering the channels.
However, alternative methods to create such chambers are also available. Please read Note 1 for a more detailed explanation.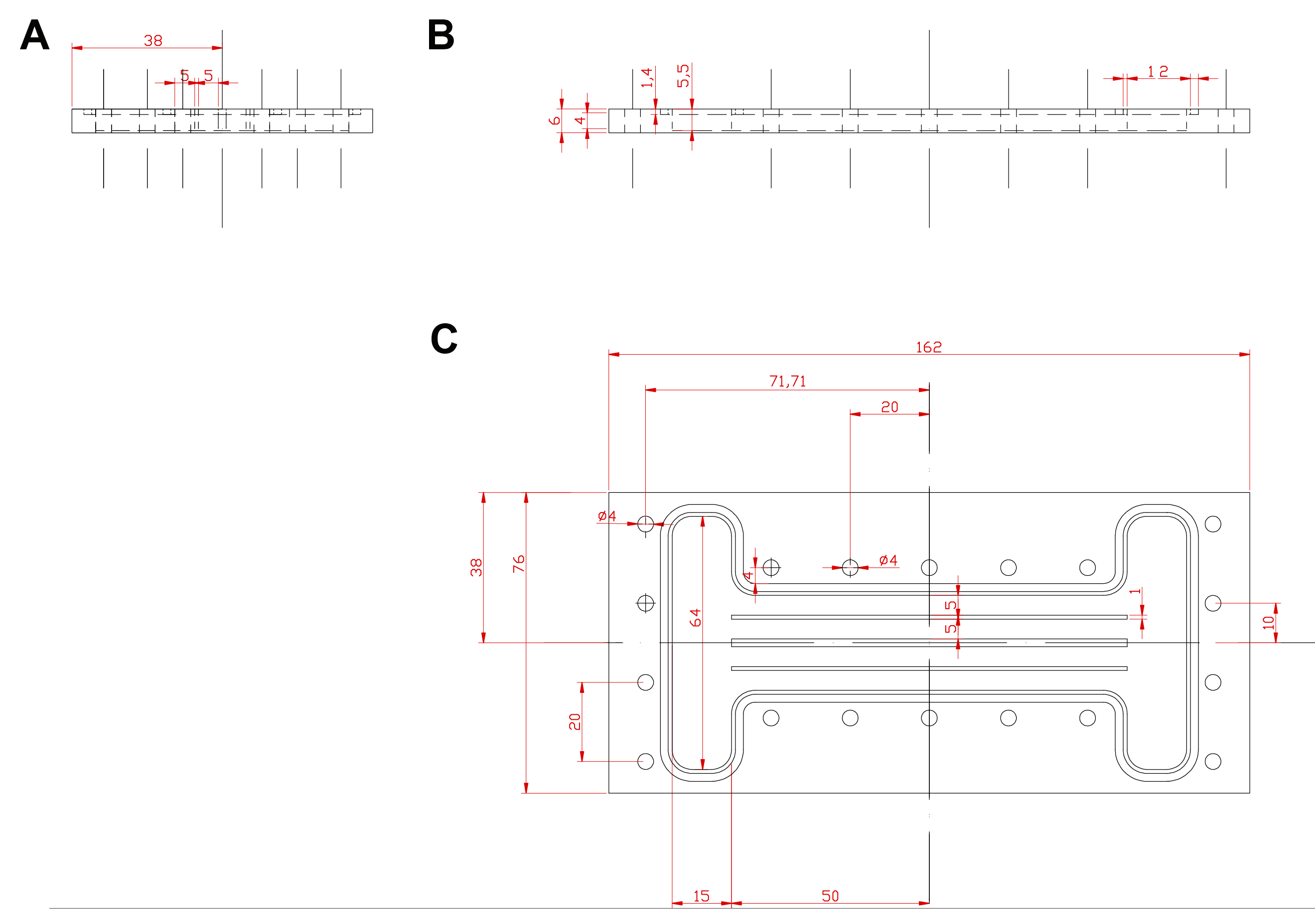
Figure 1. Custom-made biofilm chamber blueprint showing (A) side, (B) front and (C) top views. A polycarbonate sheet was carved using a milling machine following this blueprint. The current state-of-the-art in manufacturing technology allows the use of other techniques such as 3D printing to create this design or any other conceptualization in a different type of materials (i.e., PP, PC, etc.) aligned to the purpose of the experiment. Dimensions expressed in mm. - Assembly and setting up of the fluidic circuit
The fluidic circuit was designed to work at steady state regime and room temperature while maintaining a constant flow rate passing through the biofilm chamber. Losses of humidity due to constant bubbling can be minimized by using a humidity chamber to increase the relative humidity of incoming air flow. Additionally, a redundancy criteria was applied when designing the fluidic circuit for culture aeration (by using mechanical stirring and bubbling) and liquid pumping (use of two pumps in series) to avoid an eventual insufficiency in oxygen demand and increased head loss due to growing biomass within the circuit surfaces (i.e., tubing, equipment, etc.).- Organize the equipment according with the distribution depicted in Figure 2 (up) taking into account the available space in the room where the device will be placed. Figure 2 (down) shows an oblique and top view of an assembled device. By doing this, you will realize how much tube is needed to connect the different elements and how to place the different elements to make easy all the assembly, operation, inspection and maintenance tasks.
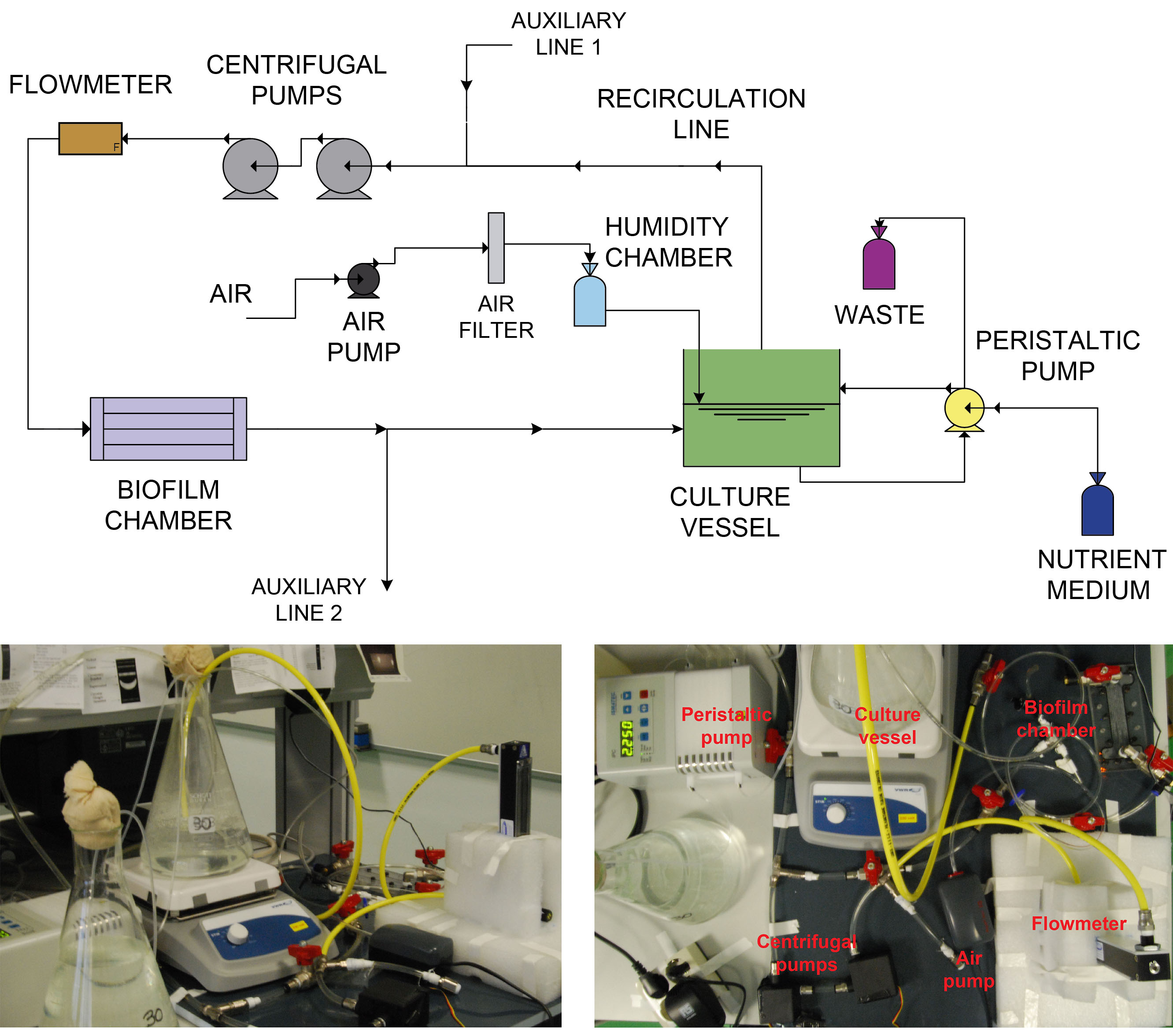
Figure 2. Schematic layout (up) and real views (down) of the experimental device. The system consists of a culture vessel containing a large volume of planktonic bacteria, a fresh input of nutrient and air to feed and aerate the culture, a waste line to work in steady state and a recirculating line to transport culture to the biofilm chamber. This recirculating line was designed to allow the flow of large amounts of liquid culture through the biofilm chamber without consuming unaffordable amount of nutrient medium. - Screw tube connectors with valves, flowmeter and flow cell as indicated in Figure 3 using PTFE tape in male threads. Tight firmly the threads without breaking the tape. Then, cut pieces of tubing with the desired length and connect them as illustrated.
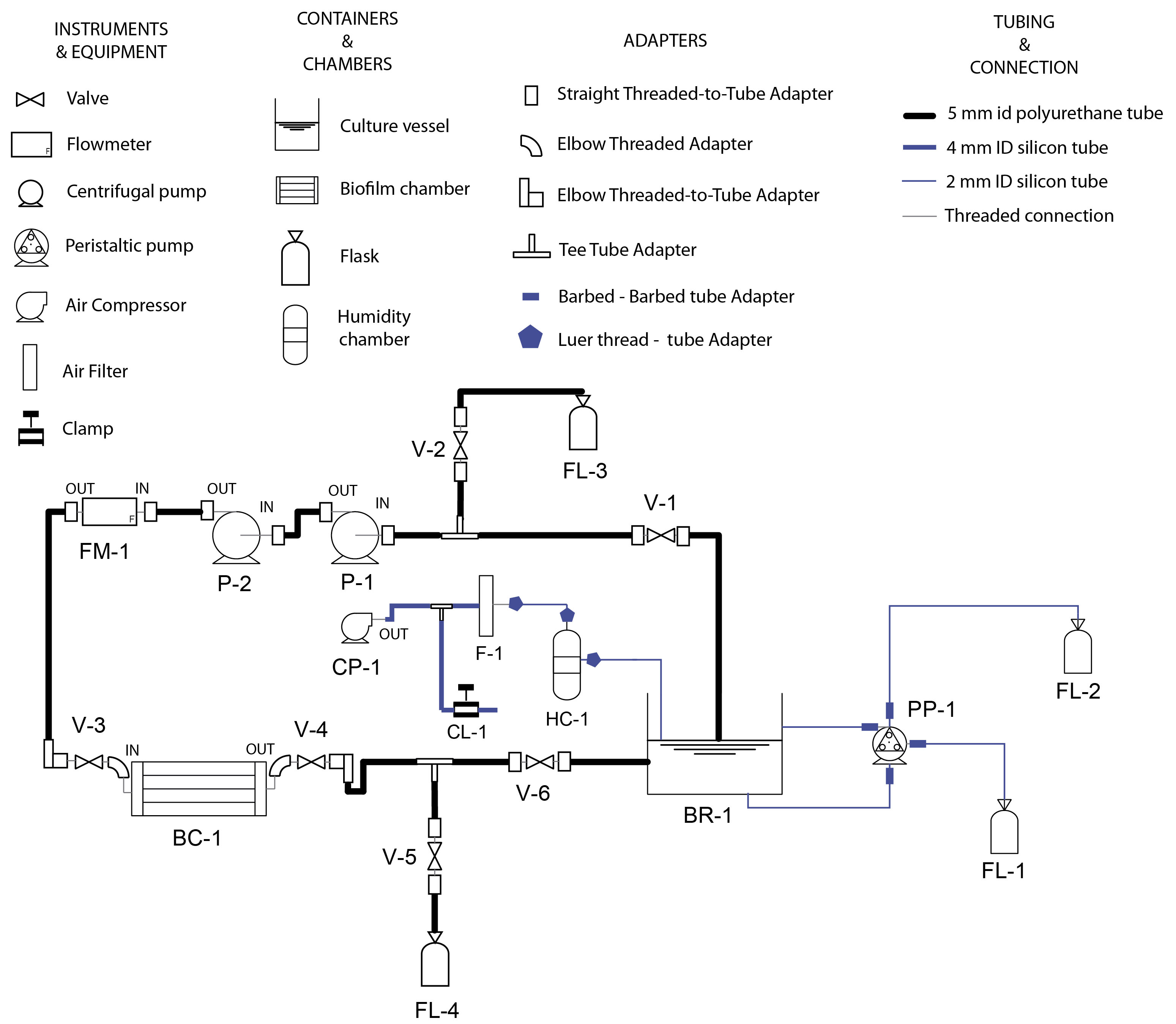
Figure 3. Assembly scheme. The device was assembled following this scheme using the different components described in the protocol. Equipment and containers were labeled following an alphanumeric coding (i.e., BR-1) to help providing detailed instructions at element level of how to manipulate the device during the start-up and operation. - Assemble the custom O-Ring to use for sealing the biofilm chamber lid: cut 400 mm of the 2 mm nitrile rubber O-Ring cord and join both ends using a drop of cyanoacrylate glue. Wait at least 15 min to let the glue being completely dry.
- Cover the flow cell/biofilm chamber with the lid and seal it using the most appropriate system (see Note 3 for more details).
- Check that all pumps and air compressor are connected according with designed flow direction in the circuit, and label the biofilm chamber inflow and outflow parts (i.e., a mark on the lid with a pen, a sticker, etc.) to have a reference and know the direction of the flow inside of it (because of the geometric symmetry of the chamber).
- Prepare electric connections of centrifugal pumps. Manually connect the centrifugal pumps and the AC/DC connectors. To do this, check that AC/DC converters are not connected to the electric grid and cut the extreme of the wire to let it free. Separate positive and negative wires and peel their extremes to allow the connection between AC/DC converter and the pump. Link together the wires of the same polarity coming from the centrifugal pump and the converter, twisting them. Cover the peeled part with electrically isolated tape. Alternatively, if the user has a crimp tool (24 AWG), a butt wire splice connector can be used to connect electrical wiring.
- Connect all electric equipment to the socket extension lead. All electric wiring must be safely placed out of water contact, thus check that wires are in an elevated position. Use any method available (tape to hang them, elevated supports to separate them from the surface where water) to avoid potential contact with any liquid in case of leaking or accidental spill.
- It is recommended to perform a leaking test to detect bad/poor/failed connections:
- Start by closing all valves of the assembled circuit.
- Fill a 2 L Erlenmeyer flask with distilled water: this would work as culture vessel (labeled as BR-1 in Figure 3) for the whole experiment. Open valve V-1 and prime the centrifugal pumps (P-1, P-2) by manually filling with distilled water the tube connecting them with the culture vessel (BR-1) to purge as much air as possible. This last step will not be necessary if peristaltic pumps are used to pump the water.
- Open the flowmeter FM-1 at maximum capacity (if present) and all the valves in the recirculation line (V-1, V-3, V-4, V-6). Switch on the pumps P-1, P-2, checking that the liquid fills the circuit completely.
- Let the liquid flow for a minimum period of time of 30 min and check periodically that there are no leaks by inspecting the presence of small drops of water in thread connectors or water spills below any element of the circuit. If a leak is detected, stop the flow and try first to tight the connection, dry the zone and restart the flow to see if water still leaks. If the leaking persists, switch off the current, purge the water through the auxiliary line 2 (see Figure 2) and dismount the affected part. Disconnect screws and replace PTFE tape if broken. Add additional layers of PTFE tape to increase isolation when required.
- Once the leaking test is ended, close valves V-1, V-3, V-4 and V-6, and manually empty culture vessel.
- Organize the equipment according with the distribution depicted in Figure 2 (up) taking into account the available space in the room where the device will be placed. Figure 2 (down) shows an oblique and top view of an assembled device. By doing this, you will realize how much tube is needed to connect the different elements and how to place the different elements to make easy all the assembly, operation, inspection and maintenance tasks.
- Initial bacterial culture and stocks
- One day before the start of the experiment (Day 0), inoculate a 50 ml Erlenmeyer flask with 10 ml of M9 minimal medium supplemented with 0.2% (w/v) glycerol as carbon source and 0.5 μg/ml of gentamycin. Add directly from a -80 °C frozen glycerol stock of P. putida mt-2.
- Incubate overnight at 30 °C in an orbital shaker set at 170 rpm.
- On the day of the experiment start (Day 1), measure the OD600 of the culture and dilute it in 1 L of fresh M9 + 0.2% (w/v) glycerol to reach an OD600 of 0.0025.
- Prepare a sterile stock of 4 L of M9 + 0.2% (w/v) glycerol supplemented with gentamycin (0.5 μg/ml) prior to start the device.
- Starting-up
- The fluidic circuit must be sterilized prior to beginning the experiment. In order to do that:
- Connect ISMATEC tubing corresponding to nutrient/waste peristaltic pumping lines PP-1 with 2 mm silicone tubing parts using barbed–barbed straight connectors. Connect also the 2 mm tube line through which the air will be conducted to the main flask with a Male Luer to 1/8" L Barb Adapter. Use aluminum foil to cover the extremes of both tube lines separately, and autoclave them.
- Fill 500 ml of 1 L Erlenmeyer flask FL-3, and 700 ml of BR-1 vessel with ethanol 70% (v/v). Introduce the magnetic stirring bar in BR-1 and connect FL-3 to auxiliary line 1. Check that valves V2, V-3, V-4 and V-6 in the recirculating loop are opened and V-1 and V-5 are closed, leaving also the flowmeter open at maximum capacity and switch on the centrifugal pumps. Transfer the stored ethanol in FL-3 to BR-1 until most of the flask is empty and then switch off pumps P-1, P-2. This will fill the auxiliary line 1 with ethanol. Close V-2 and open V-1. Then switch on again pumps P-1 and P-2, enabling the accomplishment of the sterilization of the system along 60 min.
- Stop centrifugal pumps P-1, P-2, open valve V-5, close V-6 and switch on P-1 and P-2 to let the ethanol leave the system. The removal of the ethanol has to be performed by letting the pumps to empty the culture vessel BR-1 as much as possible. Avoid its total empty while pumps are still working, as this would introduce air in recirculating line and cause priming problems in the pumps.
- Disconnect P-1 and P-2 once BR-1 is totally empty (and no air has still entered in the circuit, as said above), and replace the 1 L Erlenmeyer flask FL-3 by another one filled with 1 L of sterile water. Connect again P-1, P-2 to wash with approximately 100 ml of water the remaining ethanol within recirculating line. Close V-5 and open V-6 to let the fresh water enter in the main tank and wash the ethanol during 1 h.
- Repeat the previously described operations for ethanol–water replacement, but using 1.2 L of fresh M9 medium supplemented with 0.2% (w/v) glycerol to replace water in the system, finally leaving 1 L of fresh medium inside BR-1.
- Microscope settings will be adjusted at this point. Choose a 12-bit or 16-bit resolution for images to obtain a larger sensitivity in grayscale values. Select the channel to detect the fluorescent signal generated by the studied strain (here we recommend GFP because of its stability and performance). Take 10 μl of saturated overnight culture and spread it onto a microscope slide with the tip of the pipette. Place a cover slip onto the sample and examine it with the microscope. Adjust the gain of the microscope and laser intensity to obtain a non-saturated image. Keep these settings, as they will be constant through the experiment
- Switch off pumps P-1, P-2 and PP-1, close all valves, disassemble biofilm chamber BC-1 and place it under the microscope following the steps detailed in the image acquisition section. Gather images in the biofilm chamber (if any signal was detected, it would be caused by material and/or liquid medium self-fluorescence). These images will be used to set the background noise threshold.
- Remove aluminum foil and connect nutrient stock FL-1 and waste tank FL-2 with peristaltic pump PP-1, and their respective outputs with BR-1. Avoid any contact of the tubing with any surface when placing them inside the flasks to prevent contamination. Place the ISMATEC stop coded tubes in the pump cassettes and check that they are properly secured.
- Connect a 4 mm silicone tubing piece with the output exit of the air pump. Add a tee and connect in the 90° branch a silicone tube with a clamp CL-1, which will serve to regulate air inflow. Then connect the syringe filter by its Luer side with humidity chamber HC-1. Finally, connect the output tube of HC-1 with BR-1 by introducing the free side of the tube in the flask. See Note 2 for additional comments.
- Switch on the magnetic stirrer at 150 rpm, and connect air pump. With clamp CL-1 regulate the flow of air entering in BR-1 to avoid an excessive foam formation.
- Connect peristaltic pump at desired target flow rate.
- The fluidic circuit must be sterilized prior to beginning the experiment. In order to do that:
- Operation of the system
- Check daily that nutrient stock is not empty. Refill it manually as necessary within a sterile environment.
- The spread of biofilms through the inner surface of the recirculating line (both tubing and equipment) increases the hydrodynamic resistance to overcome, and may partially generate bottlenecks by partial clogging phenomenon. This can generate drops of flow rate in the recirculating line despite the fact flowmeter is set at a desired flow rate at the beginning of the experiment. Check daily that the flow rate value given by the flowmeter FM-1 is the chosen one for the experiment and adjust it as necessary.
- Look for any leak that could appear during the operation due to loose connections. Carefully inspect unexpected spills or liquid drops close to joints. If any, stop the pumps and tighten the joints using the adjustable spanner. Wet the leak with ethanol 70% (v/v), dry it and reconnect the system to check that the issue is fixed.
- Once the experiment is finished, replace the culture with a mixture of sterile water and sodium hypochlorite (NaClO at 10% [w/v]) using auxiliary lines 1 and 2 as described previously. Let the system recirculate the bleach for 1 h to sterilize the recirculating loop. Purge the circuit and refill it with distilled water during 1 h to wash the bleach of the system.
- Disassembly the circuit, dispose of tubing and biofilm chamber, and store spare parts and pumps.
- Repeat all previous steps for every biological replica and every tested experimental condition.
- Image acquisition process
- At the desired time, stop centrifugal pumps P-1, P-2 and peristaltic pumps PP-1, close valves V-1, V-6 and open valves V-2, V-5. Under sterile conditions, prepare 300 ml of fresh M9 medium + 0.2% (w/v) glycerol in a 500 ml Erlenmeyer flask and connect it in FL-3. Reduce the set flow rate of FM-1 by 50% of the current value and switch on the centrifugal pumps P-1, P-2. This will wash the flow cell chamber with clear nutrient medium, reducing the unspecific bright produced by planktonic bacteria when gathering images. Switch off P-1, P-2 when 400 ml of the liquid has been introduced, avoiding the entrance of air in the circuit, and close valves V-2, V-5.
- Close the valves V-3 and V-4 and disconnect them from the circuit using the adjustable spanner. Fill a beaker with ethanol 70% (v/v) and immerse both elbow threaded-to-tube adapters to avoid outer contamination. If the biofilm chamber is placed on the microscope at the beginning of the experiment, skip this step.
- Place the biofilm chamber under the microscope and use the bright field to locate the bottom of the material by looking its irregularities. If the chamber material is smooth and transparent, it can be difficult focusing the bottom of the sample (there is no visual reference to guide the user about where to find the position of the bottom): make a small spot with a marker pen at the outer face of the chamber to create a visual reference to follow with the microscope when inspecting the sample.
- Images can be taken by picking random snapshots or by performing composed images from merging images spatially distributed through a regular grid of n x m images (i.e., see Figure 4). Choose the region of the biofilm chamber to gather it and spot it with a marker if an evolution of the region is necessary. Otherwise, choose a random point in the chamber.
- Once the images have been acquired, reconnect the biofilm chamber to its original position, checking that the orientation of the chamber matches with flow direction, reset the initial flow rate in FL-1 and reconnect the device to continue the experiment (when applies).
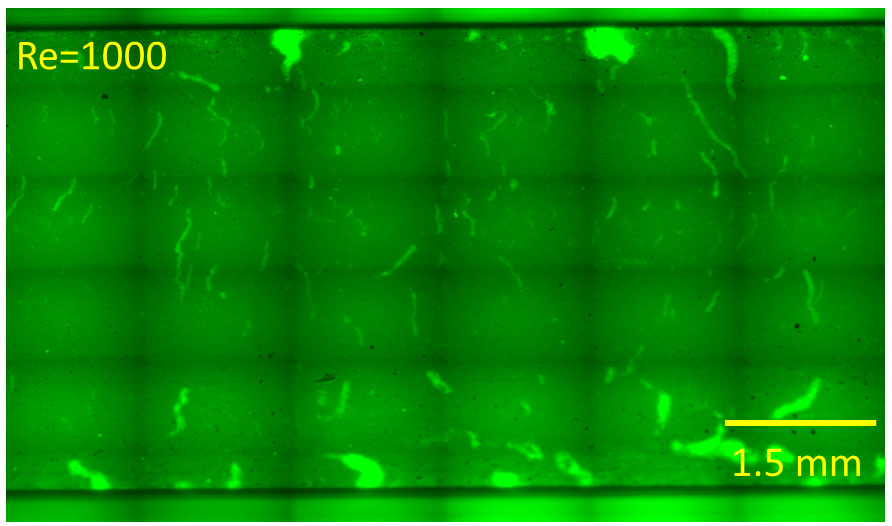
Figure 4. Tile-scan image showing a section of a custom-made chamber. The shape of the device and the whole biofilm pattern colonizing it can be observed only after merging a set of pictures forming a regular grid covering the whole width of the channel.
Data analysis
Once the experiment is finished, a set of images ![]() showing the studied biofilm morphology will be available for analysis, where k, q subscripts and t superscript indicates the k-th image of the experimental set q taken at time t. Variable names in bold style refer to vector type variable, and regular style refers to scalar quantities. Each image
showing the studied biofilm morphology will be available for analysis, where k, q subscripts and t superscript indicates the k-th image of the experimental set q taken at time t. Variable names in bold style refer to vector type variable, and regular style refers to scalar quantities. Each image ![]() will be treated as an n x n matrix in which operations of subtraction, counting and averaging will be performed for every element of these matrices. Every pixel contained in
will be treated as an n x n matrix in which operations of subtraction, counting and averaging will be performed for every element of these matrices. Every pixel contained in ![]() reports a GFP fluorescence associated intensity value captured by the microscope in a certain spatial position. Larger values of intensity are thus directly correlated with larger amounts of detected GFP in every pixel.
reports a GFP fluorescence associated intensity value captured by the microscope in a certain spatial position. Larger values of intensity are thus directly correlated with larger amounts of detected GFP in every pixel.
The procedure first treats every image to remove noise and extract relevant variables containing information about the morphology of the biofilm (see Figure 5), which will be further exploited to gather quantitative statistic parameters for analysis. Here is described how to proceed:
- Using microscope software or any other image software (i.e., ImageJ), save the images as *.tiff files.
- Convert *.tiff files to grayscale images using the chosen image analysis software.
- For every set of experimental conditions q, open the t = 0 image (
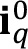 ) and compute the average (“AVG” operator) of intensity detected in the image, which will be the background signal of the biofilm chamber and nutrient medium. Round the result to the nearest integer (“round” operator) to obtain an integer to avoid decimal numbers when discounting its effect from intensity profiles.
) and compute the average (“AVG” operator) of intensity detected in the image, which will be the background signal of the biofilm chamber and nutrient medium. Round the result to the nearest integer (“round” operator) to obtain an integer to avoid decimal numbers when discounting its effect from intensity profiles.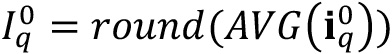
- For each available image
 , do the following loop:
, do the following loop:- Remove the autofluorescent contribution of the sample by subtracting the auto-fluorescent value
 (computed from t = 0 sample) to all pixels of the image.
(computed from t = 0 sample) to all pixels of the image.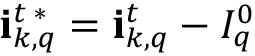
- Assign a value of 0 to all the pixels with negative value after the subtraction.
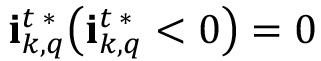
- Compute the basal part
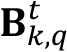 of the image, which is defined as a matrix of the same size that the processed image whose values are all equal to the average of the intensity value registered in all the pixel images (
of the image, which is defined as a matrix of the same size that the processed image whose values are all equal to the average of the intensity value registered in all the pixel images (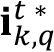 ). Conceptually it would be the fraction of the intensity of constant value detected in the image related to the existence of a homogeneous biofilm layer at the bottom of the chamber and the accumulation of GFP within biofilm cells.
). Conceptually it would be the fraction of the intensity of constant value detected in the image related to the existence of a homogeneous biofilm layer at the bottom of the chamber and the accumulation of GFP within biofilm cells. 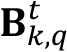 is computed by first averaging the intensity value of all the pixels in the image
is computed by first averaging the intensity value of all the pixels in the image 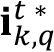 and multiplying the obtained scalar value by an n x n matrix filled with ones.
and multiplying the obtained scalar value by an n x n matrix filled with ones. 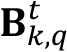 is thus a matrix with a homogeneous signal value equal to AVG(
is thus a matrix with a homogeneous signal value equal to AVG(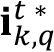 ).
).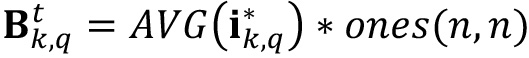
- Filter the noise value produced by GFP accumulation within cells and the presence of planktonic cells within biofilm pores. Here, different criteria can be applied depending on the user’s preference. In our case, noise is computed as:
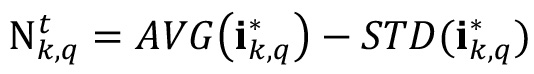
where, STD() operator computes the standard deviation of the intensity values present in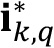 .
. - The difference between basal layer signal and noise contribution will be the estimation of the real biofilm signal covering the image (
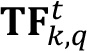 ).
).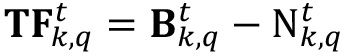
- Drop to zero all negative values of
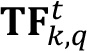 .
.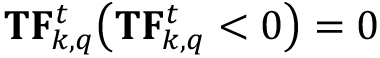
- Now, compute the fluctuating part;
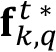 of the signal by discounting the basal signal from the image
of the signal by discounting the basal signal from the image 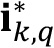 , and again drop to zero all negative values.
, and again drop to zero all negative values.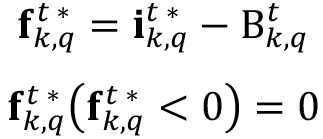
- Remove the autofluorescent contribution of the sample by subtracting the auto-fluorescent value
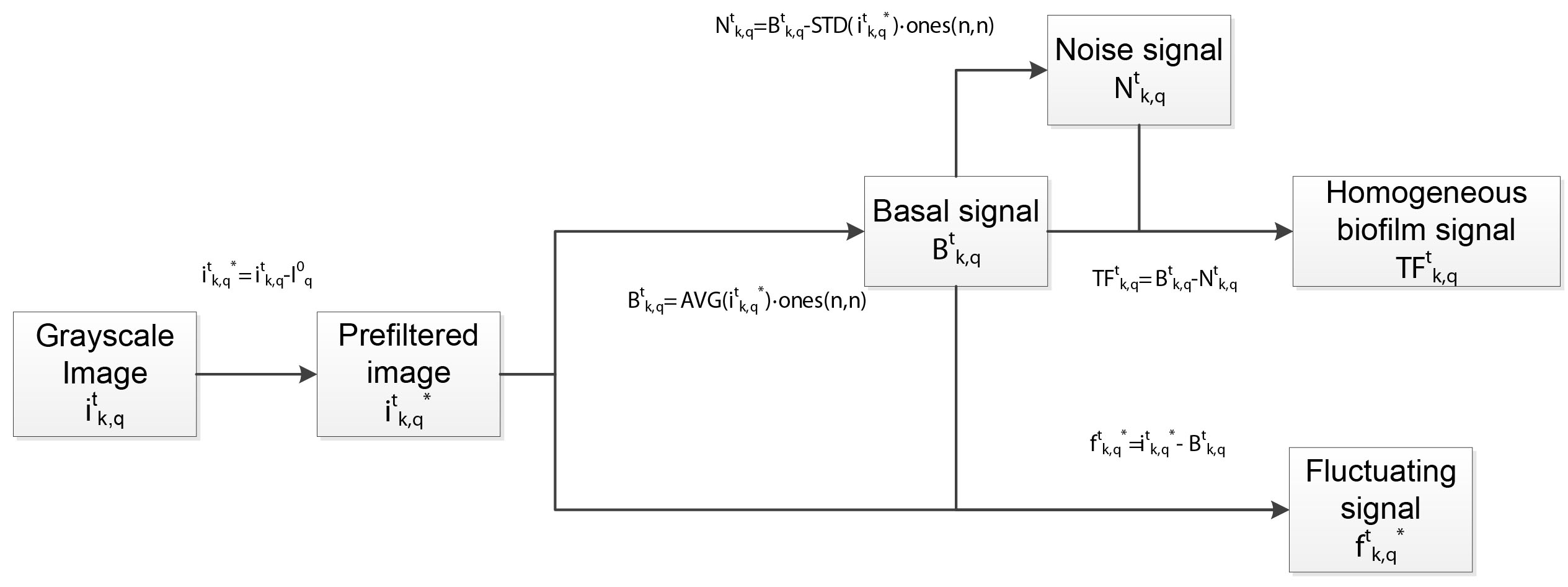
Figure 5. Filtering and morphological data extraction of grayscale images. Box diagram showing the calculation scheme to extract relevant morphological parameters (![]() and
and ![]() variables) for further statistical analysis.
variables) for further statistical analysis.
- Copy
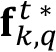 into a new variable called
into a new variable called 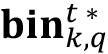 , in which a binary mask will be created by dropping to zero all values smaller than a chosen threshold th, and setting a value of one otherwise. Choose a threshold value equal to 1.
, in which a binary mask will be created by dropping to zero all values smaller than a chosen threshold th, and setting a value of one otherwise. Choose a threshold value equal to 1.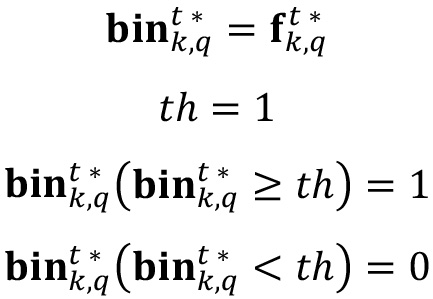
- Label the connected pixels in the binary image using a labeling algorithm included in image processing software, and calculate the area in pixels of every connected region detected in label using a regionprops-like algorithm. Note that the result of this operator is a list with different measurements of detected bodies, and then it is required to select the ‘area’ property. See Note 4 for additional information.
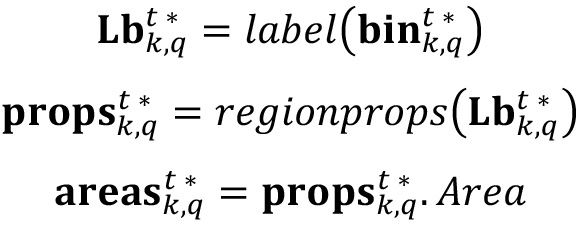
- Compute the parameters of interest just by performing selection and counting of labeled bodies. Here four metrics are evaluated: Average intensity of biofilm layer (p1), average cluster size (p2), average percent area occupied by clusters (p3) and number of clusters (p4).
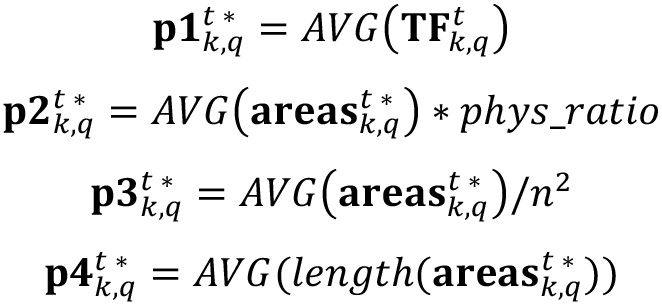
where phys_ratio is the factor employed to convert a square pixel unit to μm2, which is given once an objective and a zoom is chosen when gathering images with the microscope, the factor n2 is the maximum possible area in pixels to be covered in an image (the total number of pixels forming the image), and the length() operator provides the number of elements of a given list. - For any picture of interest, compute the cluster size distribution in pixels (
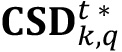 ) of labeled objects base of their occupied area. To do this, get the unique values of areas appearing in the area distribution in pixels (
) of labeled objects base of their occupied area. To do this, get the unique values of areas appearing in the area distribution in pixels ( ) and micron2 (
) and micron2 ( ) and then iterate through a loop to count the number of labeled bodies that have those area values. Inside this loop, row positions are stored in elems by filtering with a comparison operation (step 1), then the labels assigned for those positions in
) and then iterate through a loop to count the number of labeled bodies that have those area values. Inside this loop, row positions are stored in elems by filtering with a comparison operation (step 1), then the labels assigned for those positions in 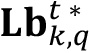 matrix are gathered from
matrix are gathered from  list (step 2) and a loop is performed to calculate averages of intensity signals for every labeled element, storing each value in an auxiliary vector aux (step 3). At the end of the loop, an average of the computed intensity values (step 4) is taken as central value of intensity for all labeled elements of
list (step 2) and a loop is performed to calculate averages of intensity signals for every labeled element, storing each value in an auxiliary vector aux (step 3). At the end of the loop, an average of the computed intensity values (step 4) is taken as central value of intensity for all labeled elements of 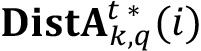 . Additionally, the number of elements of such area is stored in
. Additionally, the number of elements of such area is stored in 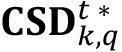 (i) to generate the Cluster Size distribution (step 5):
(i) to generate the Cluster Size distribution (step 5): - The above data calculation scheme can be applied for the whole image dataset covering all tested experimental condition range. Average and standard deviation parameters can be derived from analyzing every batch of images (having k images collected from different biological replica) for every tested experimental condition q. As a result, bar diagrams (see Figure 6A) showing averages and error bars of these batches of images, and distribution curves for sizes and other variables of interest (see Figure 6B) can be obtained. For more details, see a complete set of results in Espeso et al. (2018).
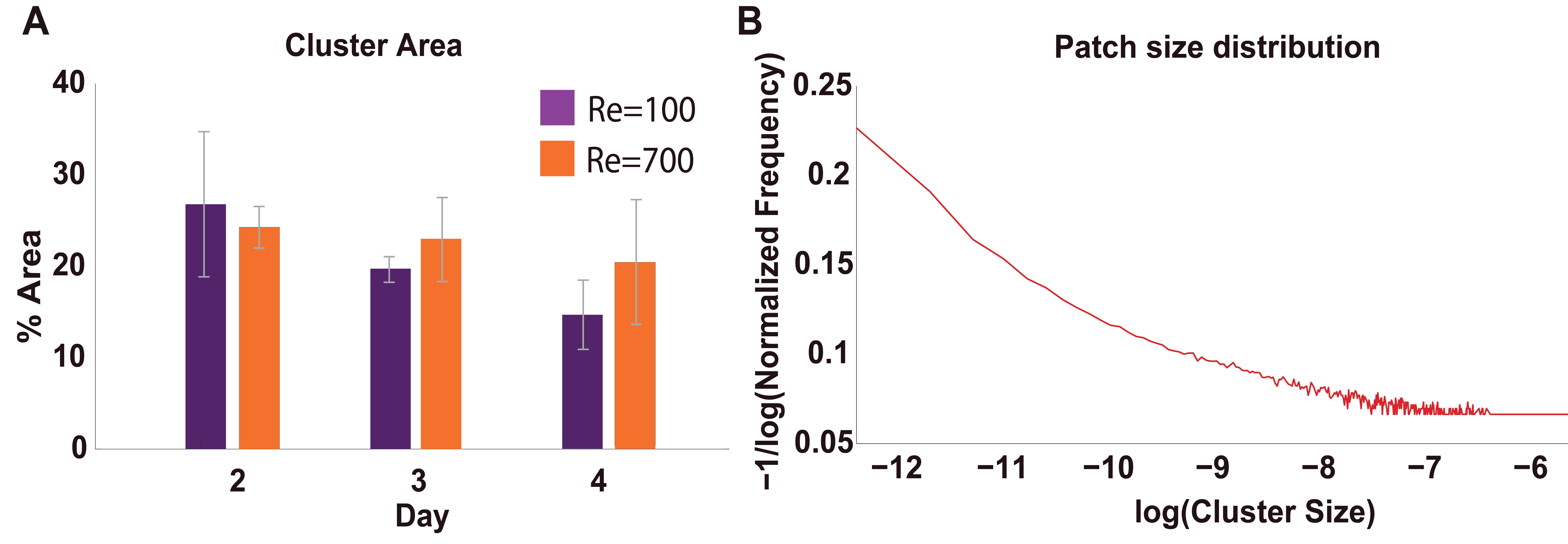
Figure 6. Typical bar diagram (A) and distribution curves (B) obtained after performing the data analysis step
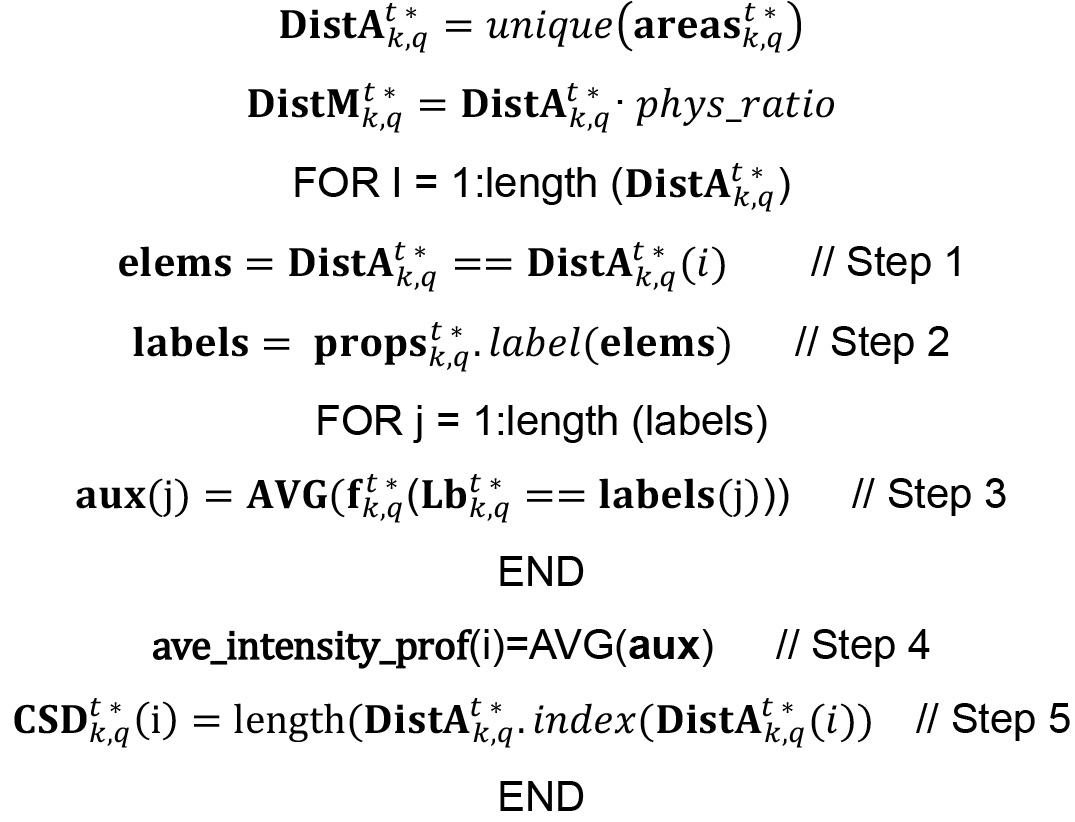
where unique () operator returns the values of a list or vector excluding repeated elements and DistA.index(x) operator provide the vector positions in the list DistA where values contained in it are equal to target value x.
With these variables, the normalized averaged intensity and the percentage of intensity for all given cluster size distribution can be derived just dividing ave_intensity_prof by the maximum or the sum of the distribution, respectively.
Notes
- When developing this protocol, we found several issues related to microscope inspection. Specifically, the use of a microscope equipped with a galvanometric tray for high sensible displacement constrains the maximum amount of weight to operate on it. The described configuration exceeded such parameters, operating out of security thresholds, which caused several problems of stability. To avoid any risk of damage for the microscope, the user should consider using an alternative approach to reduce the weight of the biofilm chamber. We suggest the replacement of cast iron valves coupled to the flow chamber by plastic valves (PVC, PP), separate valves from biofilm chamber by using flexible tubing that allow supporting valve weight out of galvanometric tray or reducing the size of the whole plate to reduce the overall material weight. Additionally, the availability of user-friendly 3D-printing technology offers the possibility to create custom made chambers without the need of using an industrial milling machine, as it was used here. Manufacturing the model with a water-resistant material (i.e., polycarbonate, polypropylene), further treatment with an organic vapor to seal material pores at microscopic level (see Tsuda et al., 2015 for details) and finally covering it with two transparent covers of the same material may be a good alternative.
- Air flow should be complemented with a humidity chamber between the air filter and the entrance of the auxiliary tank to avoid loss of liquid by desiccation. To do this, just use a two or four entrance GL-45 cap with a regular 1 L flask filled with 500 ml of autoclaved water, allowing incoming air to bubble through the water column prior to entering into the system. Connect it to the system by simply adding two pieces of silicon tubing ended with Male and female Luer to 1/8" L barb adapter (sterilization will be performed by autoclaving all together).
- In the case of small flow cells, we recommend silicon, however for larger biofilm chambers with larger flow rates a stronger fastening would be required (i.e., o-rings combined with bolts and nuts for larger chambers).
- The “regionprops” and “Label” operators here mentioned are functions implemented in image software packages (such as MATLAB or Python) that applies algorithms for labeling detected regions in images considered as “connected”, and measure typical parameters of interest (i.e., centroid, moments, etc.). Depending on the chosen programming language, invoking such functions and provided outputs may vary. Detailed information of the algorithms used by these functions may be found in Haralick and Shapiro, 1992; Jain et al., 1995; Fiorio and Gustedt, 1996; Wu et al., 2005.
Recipes
In order to prepare the minimal medium, prepare the following stocks separately:
- 10x M9 salts stock (for 500 ml of solution)
42.5 g Na2HPO4·2H2O
15 g KH2PO4
2.5 g NaCl
5 g NH4Cl
500 ml H2O - Glycerol 20% (w/v) (or any other desired carbon source, for 100 ml)
23.53 ml 85% (w/v) Glycerol
76.47 ml H2O - 1 M MgSO4 (for 100 ml of solution)
12.03 g MgSO4
100 ml H2O
Autoclave and store the stocks at room temperature - M9 minimal medium supplemented with 0.2% (w/v) glycerol (1 L)
100 ml M9 salts 10x
10 ml Glycerol 20% (w/v)
2 ml MgSO4 1 M
888 ml sterile H2O
Note: All ingredients must be mixed under sterile conditions (if the water is previously autoclaved), or mixed and filtered using a 0.45 μm PES filter unit to avoid contaminations. It is recommended to prepare it when needed to prevent contamination and minimize antibiotic degradation.
Acknowledgments
This work was funded by the HELIOS Project of the Spanish Ministry of Economy and Competitiveness BIO 2015-66960-C3-2-R (MINECO/FEDER); the ARISYS (ERC-2012-ADG-322797), EmPowerPutida (EU-H2020-BIOTEC-2014-2015-6335536), MADONNA (H2020-FET-OPEN-RIA-2017-1-766975), BioRoboost (H2020-NMBP-BIO-CSA-2018), and SYNBIO4FLAV (H2020-NMBP/0500) Contracts of the European Union and the S2017/BMD-3691 InGEMICS-CM funded by the Comunidad de Madrid (Spain) and the European Structural and Investment Funds.
Competing interests
Authors declare no conflict of interest.
References
- Espeso, D. R., Martinez-Garcia, E., Carpio, A. and de Lorenzo, V. (2018). Dynamics of Pseudomonas putida biofilms in an upscale experimental framework. J Ind Microbiol Biotechnol 45(10): 899-911.
- Fiorio, C. and Gustedt, J. (1996). Two linear time Union-Find strategies for image processing. Theor Comp Sci 154: 165-181.
- Haralick, R. M. and Shapiro, L. G. (1992). Computer and robot vision. Volume I. Addison-Wesley Longman Publishing Co., Inc. Boston. ISBN: 0201569434.
- Jain, R., Kasturi, R. and Schunck, B. G. (1995). Machine Vision. McGraw-Hill, Inc., ISBN: 0-07-032018-7.
- Tsuda, S., Jaffery, H., Doran, D., Hezwani, M., Robbins, P. J., Yoshida, M. and Cronin, L. (2015). Customizable 3D printed 'plug and play' millifluidic devices for programmable fluidics. PLoS One 10(11): e0141640.
- Wu, K., Otoo, E.J., and Shoshani, A. (2005). Optimizing connected component labeling algorithms. Medical Imaging: Image Processing.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Espeso, D. R., Martínez-García, E. and Lorenzo, V. D. (2019). Assembly of a Custom-made Device to Study Spreading Patterns of Pseudomonas putida Biofilms. Bio-protocol 9(10): e3238. DOI: 10.21769/BioProtoc.3238.
Category
Microbiology > Microbial biofilm > Biofilm culture
Microbiology > Community analysis > Spatial interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










