- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An Improved Bioassay to Study Arabidopsis Induced Systemic Resistance (ISR) Against Bacterial Pathogens and Insect Pests
(*contributed equally to this work) Published: Vol 9, Iss 10, May 20, 2019 DOI: 10.21769/BioProtoc.3236 Views: 8018
Reviewed by: Zhaohui LiuPalaniappan SivasankarHimanshu Sharma

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantitative Estimation of Auxin, Siderophore, and Hydrogen Cyanide Production in Halo and Drought-Tolerant Bacterial Isolates for Cucumber Growth
Zeinab Fotoohiyan and Ali Salehi Sardoei
Oct 5, 2025 1342 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1655 Views

Reproducible Emu-Based Workflow for High-Fidelity Soil and Plant Microbiome Profiling on HPC Clusters
Henrique M. Dias [...] Christopher Graham
Jan 20, 2026 404 Views
Abstract
The plant immune system is essential for plants to perceive and defend against bacterial, fungal and insect pests and pathogens. Induced systemic resistance (ISR) is a systemic immune response that occurs upon root colonization by beneficial microbes. A well-studied model for ISR is the association of specific beneficial strains of Pseudomonas spp. with the reference plant Arabidopsis thaliana. Here, we describe a robust, increased throughput, bioassay to study ISR against the bacterial pathogen Pseudomonas cannabina pv. alisalensis (formerly called Pseudomonas syringae pv. maculicola) strain ES4326 and the herbivore Trichoplusia ni by inoculating Pseudomonas simiae strain WCS417 (formerly called Pseudomonas fluorescens WCS417) on Arabidopsis plants grown in Jiffy-7® peat pellets. While most commonly used for Pseudomonas-triggered ISR on Arabidopsis, this assay is effective for diverse rhizosphere bacterial strains, plant species, pathogens and herbivores.
Keywords: Induced Systemic Resistance (ISR)Background
After plant infection or colonization by pathogenic or commensal microbes, a systemic defense response can ensue involving immunological “memory” or “priming” (Parker, 2009; Fu and Dong, 2013; Martinez-Medina et al., 2016). Depending on the microbe and the associated plant organ, different systemic resistance programs are induced (Pieterse et al., 2014). When roots encounter specific commensal microbes, induced systemic resistance (ISR) in distal root and shoot tissue is observed (Pieterse et al., 1996; van Loon et al., 1998). Although the genetic and hormonal signaling mechanisms deployed during the ISR response are partially understood (Verhagen et al., 2004; Pieterse et al., 2014; Pangesti et al., 2016), many questions still remain unanswered, such as the mechanisms involved in root-microbiome communication or the identity of systemic signal(s) (Pieterse et al., 2014; Haney et al., 2018; Cecchini et al., 2019).
The inoculation of Arabidopsis thaliana roots with Pseudomonas simiae WCS417 has been used as a model system for studying ISR against bacteria, fungi and herbivore invaders (Pieterse et al., 1996; van Loon et al., 1998; Verhagen et al., 2004; Haney et al., 2015). Existing bioassays for P. simiae-Arabidopsis ISR make use of sterile sand-germinated Arabidopsis followed by seedling transplantation into a sand/soil mixture containing the rhizobacteria (Pieterse et al., 1996; Pozo et al., 2008; van Oosten et al., 2008). Building on this method, we have recently developed ISR bioassays against the bacterial pathogen Pseudomonas cannabina pv. alisalensis (formerly called Pseudomonas syringae pv. maculicola) strain ES4326 (Pma) and the herbivore Trichoplusia ni (T. ni) using Arabidopsis plants germinated and grown in Jiffy-7® peat pellets. This is an effective system to robustly trigger and quantify ISR, primarily because peat pellets have a constant volume and low fluorescent pseudomonad content (Haney et al., 2015 and 2018; Cecchini et al., 2019). Moreover, avoiding the transplantation of seedlings reduces the experimental labor and stress to the seedlings, thereby improving assay efficiency. Here, we describe a step-by-step bioassay methodology for ISR experiments that can be potentially standardized across laboratories worldwide.
Materials and Reagents
- 96-multiwell plates (Corning, Costar, catalog number: 2797)
- Kimwipes (Fisher, catalog number: 06-666A)
- 15 ml and 50 ml centrifuge tubes (MidsciTM, catalog numbers: 15 ml-CT2715, 50 ml-CT2750)
- Plastic domes (Hummert International, catalog number: 11-33480) and trays (Hummert International, catalog number: 11-33010)
- 9 cm round Petri dishes (Fisher, catalog number: FB0875712)
- 1.5 ml microfuge tubes (MidsciTM, catalog number: AVSS1700)
- Plastic pestles for 1.5 ml microfuge tubes (Fisher, catalog number: 12-141-364)
- 3 or 5 mm metal beads (QIAGEN, catalog number: 69997 or 69989)
- Trays for holding the Jiffy-7® peat pellets (Hummert International, catalog number: 11311000)
- 1 ml-syringes without needle (BD Biosciences, catalog number: 309659)
- Mesh bags (can be made by sewing a semi-oval out of a fine washable mesh material such as bridal veil; see Figure 3 for schematic; alternatively, perforated cellophane bread bags can be used)
- Tightly closing plastic container (with screwcap lid)
- Damp paper towel
- Fine bristled paint brush
- 96-well racked collection microtubes (optional, for 96-well tissuelyser format) (QIAGEN 19560)
- Arabidopsis thaliana (L. Heyhn.) ecotype Columbia (Col-0)
- Pseudomonas cannabina pv. alisalensis (formerly called P. syringae pv. maculicola ES4326 (Bull et al., 2010) carrying an empty vector (PmaDG3/Pma) (Guttman and Greenberg, 2001) bacterial culture (stored in 15% glycerol at -80 °C)
- Trichoplusia ni (eggs; Benzon Research or Natural Resources Canada)
- Pseudomonas simiae strain WCS417 (formerly called Pseudomonas fluorescens WCS417)
- Jiffy-7® peat pellets (Jiffy products, Canada, Hummert International, catalog number: 14-23700)
- Glycerol (Sigma-Aldrich, catalog number: G7757)
- K2HPO4 (Fisher, catalog number: P288)
- MgSO4 (Fisher, catalog number: BP213)
- Proteose peptone No. 3 (BD Biosciences-US, catalog number: 211693)
- Antibiotics
- Rifampicin (GoldBio, catalog number: R-120)
- Kanamycin (GoldBio, catalog number: K-120)
- Agar (Fisher, catalog number: BP1423)
- Sterile distilled and tap water
- Bleach (Clorox Concentrated Germicidal Bleach)
- Ethanol (Decon Labs Inc., catalog number: 2701)
- Triton X-100 (Fisher, catalog number: BP151)
- Tryptone (BD Biosciences-US, catalog number: 211705)
- Sodium chloride (NaCl)
- Yeast extract (BD Biosciences-US, catalog number: 212750)
- 70% ethanol (see Recipes)
- 25% bleach supplemented with 0.1% Triton X-100 (see Recipes)
- 0.1% agar (see Recipes)
- King's medium B (KB) (see Recipes)
- Luria-Bertani medium (LB) (see Recipes)
- 10 mM MgSO4 (see Recipes)
Equipment
- Laboratory glassware
- 200 µl and 1 ml, and 20-200 µl multichannel micropipettes (Gilson)
- Forceps (Grainger, catalog number: 4CR15)
- 1 L beaker (Pyrex, 1000)
- Handheld electric drill (DeWALT, model: DWD110)
- Cork borer 4 mm diameter
- Autoclave (Primus Sterilizer Co. Inc. 1317. C.R.N: 09415.1256)
- Microcentrifuge (Eppendorf, model: 5415D)
- Freezer (-80 °C) (Panasonic VIP Plus, model: MDF-V76VC-PA)
- Plant Growth chamber at 20-23 °C with 12 h light/12 h dark (~75-100 μmol s-1m-2, cool white fluorescent; A1000 Conviron Growth Chamber with Arabidopsis Kit, or similar. Alternatively, a growth-room with ~135-145 μmol s-1m-2 -mix 50/50 of 400-watt sodium and metal halide bulbs or 75-100 μmol s-1m-2 cool white fluorescent bulbs can be used)
- Shaker incubator at 28 °C (Barnstead Max, model: Q 5000)
- Balance (Mettler Toledo, model: PB1501)
- Spectrophotometer (Bio-Mini SHIMADZU)
- Laminar flow hood (SterilGARD 3 Advance)
- Incubator at 28 °C (VWR, 3020)
- Analytical balance for weighing caterpillars
- TissueLyser Beadmill (QIAGEN, catalog number: 85300) with 24 or 96 adapters (QIAGEN, catalog number: 69982 or 69984)
Procedure
- Hydration of Jiffy-7® peat pellets and Arabidopsis seed germination and growth (Figures 1A-1C)
- In 1.5 ml Eppendorf tubes, surface-sterilize 100-200 Arabidopsis seeds by washing with 70% ethanol for 2 min followed by 5 min with 25% bleach supplemented with 0.1% Triton X-100 and three washes in sterile water. Resuspend the seeds in 1-1.5 ml of a sterile solution of 0.1% agar (prepared in water and sterilized by autoclaving) using a pipette. Alternatively, chlorine gas sterilization can be used to eliminate endophytic bacteria, particularly to avoid microbial contamination for rhizosphere microbiome studies (Haney et al., 2015).
- Prior to use, leave the seeds for 3-7 days at 4 °C to allow for their stratification.
- Prepare 8-12 Jiffy-7® pellets for each genotype per bacterial infection experiment (or 25-30 pellets per treatment for T. ni assays, see Data analysis) by placing them in a germination tray.
- Hydrate Jiffy-7® peat pellets (Jiffy Products International, Canada) by immersing the base of the pellet with tap water. Allow the pellets to stand in water for at least 1 h to allow them to hydrate.
- Once the pellets are hydrated, use a pair of clean/sterile forceps to gently even/level the top surface of the pellet.
- Sow 3-4 stratified seeds by pipetting them at the surface of each Jiffy-7® pellet and cover the trays with a plastic dome. Do not cover the seeds with soil.
- Transfer the trays to a growth chamber/room under 12 h day and 12 h night conditions at 23-20 °C day/night temperature regime, ~75-100 μmol s-1m-2 of cool white fluorescent light at rosette level, 50%-70% relative humidity. Alternatively, ~135-145 μmol s-1m-2 of mix 50/50 of sodium and metal halide light can be used.
Note: Intensities of cool white fluorescence > 125 μmol s-1m-2 cause plants to bolt between 3 and 4 weeks and so should not be used. - At 3-7 days post germination, carefully remove extra seedlings with clean/sterile forceps from the surface of the pellets leaving only a single seedling in each pellet (do not water seedlings during the first week after planting).
- Two to three times per week, bottom water the Jiffy pellets by flooding the tray with tap water and pouring the excess water off after 30-60 min.
- P. simiae suspension preparation and seedling/pellets inoculation (Figures 1D-1F)
- When seedlings are eight days old, streak P. simiae WCS417 strain from a frozen glycerol stock onto a King’s B (KB) solid medium plate supplemented with Rifampicin (100 μg/ml). Allow it to grow for 24 h at 28 °C. Pick a single colony, streak it to a new plate and grow it for another 24 h.
- Inoculate 15 ml of media supplemented with Rifampicin (100 μg/ml) (in a 50 ml Falcon tube) with a loop of confluent bacteria from the plate. Grow in a 28 °C shaker incubator for 16-24 h.
- Centrifuge the culture at 3,000 x g for 5 min, discard the supernatant and resuspend the pellet in 15 ml of sterile 10 mM MgSO4 solution. Repeat the centrifugation and finally resuspend the pellet in 15 ml 10 mM MgSO4 by pipetting up and down.
- Make a 1/10 or 1/20 dilution of the bacterial suspension (900 µl 10 mM MgSO4 + 100 µl bacterial suspension for 1:10 or 950 µl 10 mM MgSO4 + 50 µl bacterial suspension for 1:20 dilution) in a 1.5 ml tube and measure optical density at 600 nm (OD600) in a spectrophotometer to estimate the OD600 of the original P. simiae culture. (Divide the measured OD by the dilution factor).
- In a clean (non-sterile) beaker, prepare at least 1 L of the P. simiae WCS417 inoculum by diluting the bacterial suspension culture in 10 mM MgSO4 to a final OD600 of 0.01.
- Inoculate the ten-day-old seedlings with 6 ml of the bacterial solution per plant by gently pipetting on the top surface of the Jiffy-7® pellet, near the base of the seedling, taking care to avoid contact with the leaves or hypocotyl. Then, submerge the entire pellets for ~30 s in the same solution (using the 1 L beaker with the suspension) without allowing contact of the bacteria suspension with the seedling aerial tissues. One liter of bacterial solution can support the inoculation of at least 16 pellets. For the control/mock-treated plants follow the same steps but only using sterile MgSO4.
Alternatively: If multiple strains are inoculated onto plants within the same flat, to avoid cross-contamination of bacterial strains between pellets, 2 ml of more concentrated inoculum (OD600 = 0.02-0.1) can be applied to the Jiffy pellets. For these assays, the pellets should be fairly dry (generally not watered after the initial soaking) and 2 days should pass after inoculation before flood watering the plants. Using this method, multiple bacterial strains and buffer-treated controls can be grown within the same flat with no detectable cross-contamination between pellets. Additionally, significantly smaller starting volumes of inoculum can be prepared. - After these treatments, bottom water the plants as before for an additional 15-25 days (3.5-5 week-old plants) for Pma assays or 20 days (4.5 week-old plants) for T. ni assays.
Note: Different genotypes can be planted side by side in the same tray. Rotate different trays 2-3 times per week to minimize growth variation.
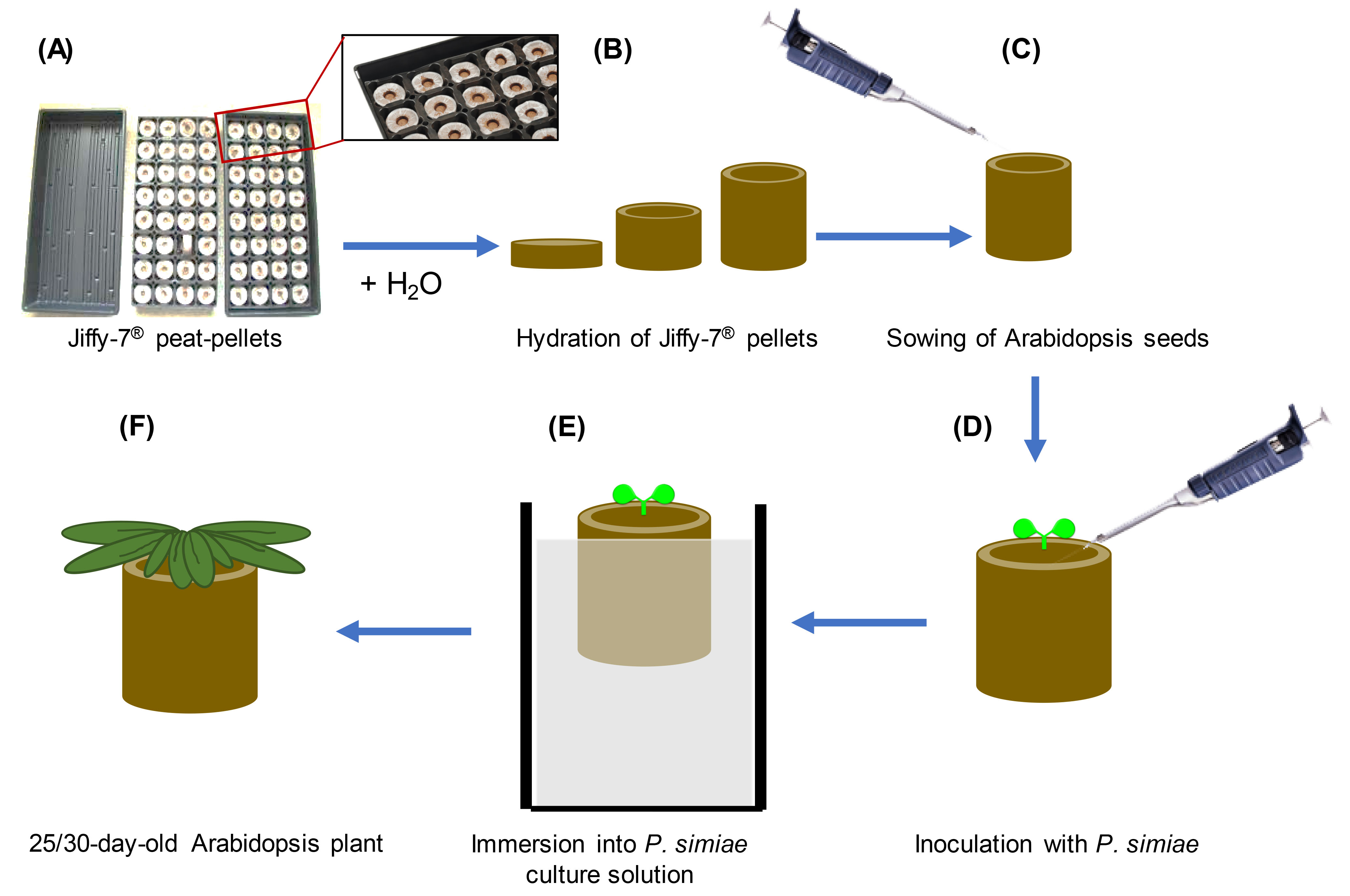
Figure 1. Schematic of P. simiae inoculation. A. Arrange Jiffy-7® peat pellets onto flats as shown. B. Flood the base of the trays with tap water for at least 60 min to allow the pellets to hydrate. C. Sow previously sterilized and stratified Arabidopsis seeds onto the surface of hydrated Jiffy® pellets. Water the pellets by flooding the base of the tray three times per week. D. Ten days after germination, inoculate 6 ml of P. simiae solution with OD600 = 0.01 onto the surface of the pellet taking care to avoid contact with the seedling aerial tissues. E. Immerse for ~30 s the Jiffy® pellet into a 1 L solution of P. simiae of OD600 = 0.01. One liter of bacterial solution can support inoculation of at least 16 pellets. F. Continue bottom-watering the pellets with tap water for a further 15 days (or 20 days for T. ni assay) until the plants are ready for plants infection with Pma or T. ni. - Infections and growth of Pma or T. ni to analyze the effects of P. simiae WCS417 ISR
To evaluate ISR against Pma (Figure 2)
Growing Pma and infection- Inoculate O/N cultures of Pseudomonas cannabina pv alisalensis carrying an empty vector (Pma) (Guttman and Greenberg, 2001) strain in KB-media supplemented with Kanamycin (50 μg/ml).
- The next morning, dilute the cultures back in KB medium supplemented with Kanamycin (1:5 ml) and grow for additional 3-4 h.
- Spin the bacteria down at 3,000 x g for 3 min and resuspend in sterile 10 mM MgSO4. Repeat the wash an additional time.
- Measure OD600 of the suspension by diluting 1:20 as described above in section B, point 4. Dilute the culture to an OD600 = 0.2 or 0.3 and dilute to a final OD600 = 0.0002-0.0003 by performing three 10-fold serial dilutions in sterile 10 mM MgSO4. The final volume should be 10 ml (~0.1 ml inoculum/leaf is required).
- Use a 1 ml syringe without a needle pressed up against the abaxial side of the 5th and 6th Arabidopsis leaves to infiltrate the bacteria into the apoplast of an entire leaf. Infect at least two leaves on eight different plants. Use a felt-tipped marker to mark the petiole of infected leaves for identification leading to the sampling and quantification next steps (see below).
Note: Inoculate plants around 1 pm. - With a Kimwipe, dry the excess bacterial suspension from each infiltrated leaf. Let the leaves dry for 1 h until the liquid in the apoplast is no longer visible. Then, cover the plants with a plastic dome to increase the humidity and reduce the variability in bacterial growth. Return plants to the growth chamber for 2-3 days.
Quantifying bacterial growth in leaf tissues- Two or three days after Pma inoculation, excise 8 leaf discs from different plants per treatment from infected leaves (ISR-induced vs. mock) using a cork borer (4 mm diameter). Take the discs from approximately the same leaf position for all the samples, 3-5 mm from the leaf tip.
- Using forceps, place each leaf disc in a 1.5-ml microfuge tube containing 200 µl of 10 mM MgSO4.
- Grind the samples using a small hand-held electric drill with a plastic pestle. Thoroughly macerate the tissue until pieces of intact leaf tissue are no longer visible to the eye (avoiding heating up the samples by performing repeated short bursts of ~2 s instead of grinding continuously for a long-time interval). We suggest grinding each sample for a similar duration to minimize variability during the experiment. Samples can also be ground with the plastic pestle by hand (without the drill).
Alternatively: Use a bead mill or TissueLyser to homogenize the samples. To use a TissueLyser, place the leaf disk into a 2 ml Eppendorf tube with a 5 mm metal bead and 100 µl water. Alternatively, a 96-well plate format can be used with 3 mm beads. Homogenize tissue for 2 min at 25 Hz. - Next, vortex the homogenate and remove 20 µl from each sample and dilute in 180 µl 10 mM MgSO4 in 96-well multi-well plate. Use a multichannel pipette to repeat this process 4 times to have a serial 1:10 dilution series (10-1 to 10-5) for each sample.
- Plate 20 µl aliquots of the 1:10 serial dilutions (10-2 to 10-5) on KB medium plates supplemented with Kanamycin (50 μg/ml of media) and allowed to dry onto the surface. Divide each KB plate with a marker such that at least eight samples/dilutions per plate can be plated.
Alternatively: By pouring media into rectangular plates, 10 µl can be spotted with a multichannel pipette, and all samples can be plated on a single plate (see Figure 2). - Incubate the plates at 28 °C for 2-3 days until colony-forming units (CFU) can be counted. Calculate the number of CFU per leaf disc by multiplying by the corresponding dilution factor (see Data analysis).
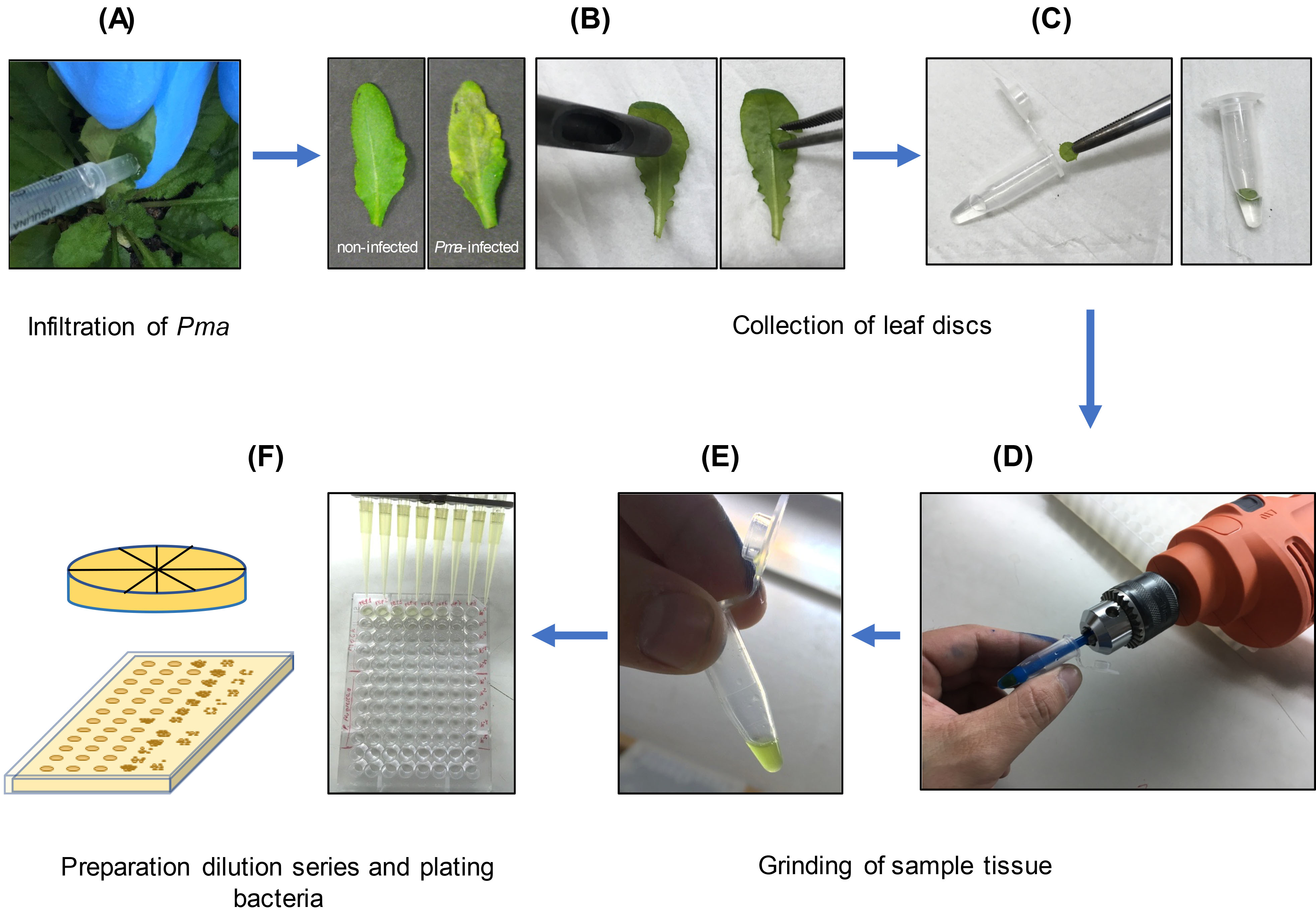
Figure 2. Images showing the key steps to evaluate ISR against Pma in Arabidopsis grown in Jiffy-7® peat pellets. A. Infiltrate the bacteria suspension into the abaxial surface of Arabidopsis leaves with a 1 ml-syringe without a needle (Pma, OD600 = 0.0003) (Note: Inoculate plants at ~1 p.m.). B. Two-three days after Pma infection, excise leaf discs from the infiltrated leaves using a cork borer (4 mm diameter). C. Place each leaf disc in a 1.5-ml microfuge tube with 200 µl of 10 mM MgSO4 solution. D. Grind the samples using a plastic pestle mounted in a hand-held electric drill (shown) or a bead beater/TissueLyser (not shown). E. Grind the leaf discs until the tissue is thoroughly homogenized and no visible pieces of tissue remain (Note: Avoid heating up the samples by performing ~2 s bursts). F. Remove 20 µl from each sample and dilute in 180 µl 10 mM MgSO4 in a multi-well plate and repeat this process to have a 1:10 dilution series (10-1 to 10-5) for each sample. Plate 20 µl aliquots of each dilution on KB medium plates or 10 µl aliquots onto rectangular plates.
To evaluate ISR against Trichoplusia ni (Figure 3)- Incubate T. ni eggs (Benzon Research or Natural Resources Canada) at 30 °C for 36 h with 12-h light days. The photoperiod of the chamber should be in synch with that of the plants so the caterpillars are entrained with the same photoperiod.
- Using a minimum of 25 plants per treatment at 4-5 weeks of age, randomly choose one newly hatched caterpillar larva from the batch. Using a small paint brush, place one larva at the center of the rosette of a Jiffy-7® pellet grown Arabidopsis rosette pretreated with Pseudomonas or buffer control.
- Cover each pellet/plant with a mesh bag (Figure 3), return to a growth chamber, and allow the caterpillars to feed for 7 days. Plants should be at a density of no more than 30 per flat.
- On the 7th day, remove the mesh bag and find the caterpillar on the plant. Weigh the larva with a precision balance to the nearest tenth of a milligram. The weight of newly hatched larvae is negligible; thus, the final caterpillar weight correlates with how much the caterpillar ate and host plant susceptibility (Cui et al., 2002).
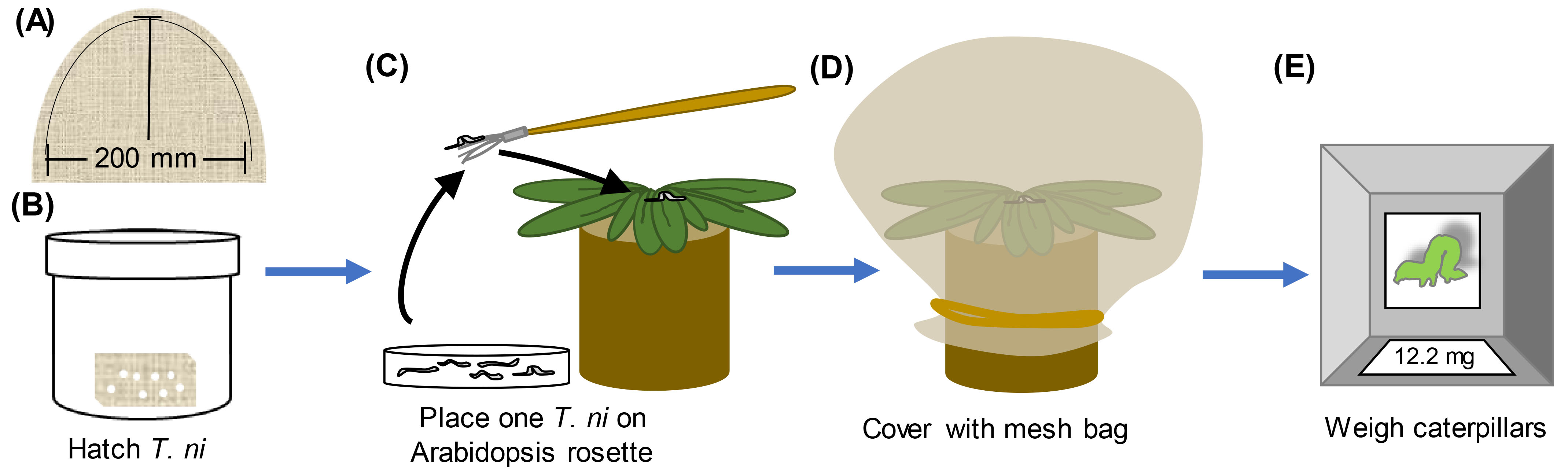
Figure 3. T. ni herbivory assay. A. Mesh bags can be made by cutting mesh (see materials) in a half-oval of ~200 x 200 mm and then using a sewing machine to sew two half-ovals together (dashed line). B. Place cheese cloth with T. ni eggs in a tightly sealing container with a damp paper towel at the bottom. Place the container in an incubator at 30 °C with the same light regime as the plants until the larvae hatch (~24-36 h). C. Using a fine paint brush, transfer a small number of newly hatch T. ni larvae to a Petri plate or other small container. Reseal the original container. Using the paint brush, place 1 larva on the center of the rosette of a 4-5 week-old Arabidopsis plant. D. Cover each plant with a mesh bag and secure at the base with a rubber band. Return the plants to the flats and return flats to the incubator. Allow caterpillars to feed for 1 week. E. One week later, find and weigh each individually (to the nearest 0.1 mg).
Data analysis
ISR-Pma data analysis
When the colonies are still small, count the CFUs for one of the dilutions (where 10-50 CFU can be counted and colonies are clearly distinct) for each disc from one leaf per plant taken and estimate the number of bacteria by multiplying with the corresponding dilution factor. Perform CFU counting for at least eight discs (12 discs provide greater statistical power and allows for increased confidence in smaller differences in bacterial growth). To increase confidence in possible differences in growth, replicate the entire experiment at least 3 independent times on different days and from different batches of inoculum. Use all the data collected from all independent experiments performed to calculate the average CFU per leaf disc plus/minus standard error. To determine if differences are statistically significant, perform analysis of variance (ANOVA) and a post-hoc test such Tukey’s HSD test or Newman-Keuls (SNK) by using appropriate statistical software (Figure 4D and methods in Cecchini et al., 2019). If only two conditions are being compared, a Student’s t-test can be used. Plot the data on a log10 scale.
ISR-T. ni data analysis
Weigh at least 25 larvae/plants per treatment, per experiment and repeat the experiment a minimum of 3 independent times. Use all the data collected from all independent experiments performed to calculate the average weight per larvae plus/minus standard error. To determine if differences are statistically significant, perform analysis of variance (ANOVA) and a post hoc test such Tukey’s HSD test or Newman-Keuls (SNK) by using appropriate statistical software. If only two conditions are being compared, a Student’s t-test can be used. If the experiments are done in different chambers or with insects from different sources, a significant replicate effect has been observed (Haney et al., 2018). As a result, data can be normalized to the buffer-treated Col-0 control data from a single experiment prior to averaging the data from at least 3 independent experiments.
Notes
Using Jiffy-7® has allowed us to study interactions between diverse below and above ground commensals and pathogens. This setup has been used to study diverse rhizosphere bacteria that induce systemic resistance or susceptibility (Haney et al., 2015 and 2018; Melnyk et al., 2019). Additionally, it can be used to study below ground effects on diverse above ground pathogens including pathogenic Pseudomonas and Xanthomonas spp. (Haney et al., 2018) and fungal pathogens such as Botrytis, and downy mildew.
Recipes
- 70% ethanol
73.7 ml of 95% ethanol
Add distilled water up to 100 ml - 25% bleach supplemented with 0.1% Triton X-100
25 ml bleach and 100 µl of Triton X-100
Add distilled water to 100 ml - 0.1% agar
Dissolve 0.1 g agar in 100 ml sterile distilled water by autoclaving, swirl the solution while it cools - King's medium B (KB)
20 g of proteose peptone No. 3
10 g glycerol
1.5 g MgSO4
1.2 g K2HPO4
For solid medium add 13 g agar
Add distilled water up to 1 L
Sterilize by autoclaving - Luria-Bertani medium (LB)
10 g tryptone
10 g sodium chloride (NaCl)
5 g yeast extract
Add distilled water up to 1 L
Sterilize by autoclaving - 10 mM MgSO4
0.12 g of MgSO4
Add distilled water to 100 ml
Sterilize by autoclaving
Acknowledgments
We thank Elizabeth Baldo and Joanna Jelenska for helping identify catalog numbers for materials and equipment.
This work was supported by NSF grant IOS1456904 to JTG, by an NSERC Discovery Grant (NSERCRGPIN-2016-04121) awarded to CHH, a China Postdoctoral Science Foundation Fellowship awarded to YS and an Agencia Nacional de Promoción Científica y Tecnológica grant (PICT-2017-0589) awarded to NMC. NMC is a Career Investigator of CONICET (Argentina).
Competing interests
The authors declare that they have no conflict of interests.
References
- Berendsen, R. L., van Verk, M. C., Stringlis, I. A., Zamioudis, C., Tommassen, J., Pieterse, C. M. and Bakker, P. A. (2015). Unearthing the genomes of plant-beneficial Pseudomonas model strains WCS358, WCS374 and WCS417. BMC Genomics 16: 539.
- Bull, C. T., Manceau, C., Lydon, J., Kong, H., Vinatzer, B. A. and Fischer-Le Saux, M. (2010). Pseudomonas cannabina pv. cannabina pv. nov., and Pseudomonas cannabina pv. alisalensis (Cintas Koike and Bull, 2000) comb. nov., are members of the emended species Pseudomonas cannabina (ex Sutic & Dowson 1959) Gardan, Shafik, Belouin, Brosch, Grimont & Grimont 1999. Syst Appl Microbiol 33(3): 105-115.
- Cecchini, N. M., Roychoudhry, S., Speed, D. J., Steffes, K., Tambe, A., Zodrow, K., Konstantinoff, K., Jung, H. W., Engle, N. L., Tschaplinski, T. J. and Greenberg, J. T. (2019). Underground azelaic acid-conferred resistance to Pseudomonas syringae in Arabidopsis. Mol Plant Microbe Interact 32(1): 86-94.
- Cui, J., Jander, G., Racki, L. R., Kim, P. D., Pierce, N. E. and Ausubel, F. M. (2002). Signals involved in Arabidopsis resistance to Trichoplusia ni caterpillars induced by virulent and avirulent strains of the phytopathogen Pseudomonas syringae. Plant Physiol 129(2): 551-564.
- Fu, Z. Q. and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839-863.
- Guttman, D. S. and Greenberg, J. T. (2001). Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol Plant Microbe Interact 14(2): 145-155.
- Haney, C. H., Samuel, B. S., Bush, J. and Ausubel, F. M. (2015). Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants 1(6): pii: 15051.
- Haney, C. H., Wiesmann, C. L., Shapiro, L. R., Melnyk, R. A., O'Sullivan, L. R., Khorasani, S., Xiao, L., Han, J., Bush, J., Carrillo, J., Pierce, N. E. and Ausubel, F. M. (2018). Rhizosphere-associated Pseudomonas induce systemic resistance to herbivores at the cost of susceptibility to bacterial pathogens. Mol Ecol 27(8): 1833-1847.
- van Loon, L. C., Bakker, P. A. H. M., and Pieterse, C. M. J. (1998). Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol 36: 453-483.
- Martinez-Medina, A., Flors, V., Heil, M., Mauch-Mani, B., Pieterse, C. M. J., Pozo, M. J., Ton, J., van Dam, N. M. and Conrath, U. (2016). Recognizing plant defense priming. Trends Plant Sci 21(10): 818-822.
- Melnyk, R. A., Beskrovnaya, P., Liu, Z., Song, Y. and Haney, C. H. (2019). Bacterially produced spermidine induces plant systemic susceptibility to pathogens. bioRxiv: 517870.
- van Oosten, V. R., Bodenhausen, N., Reymond, P., van Pelt, J. A., van Loon, L. C., Dicke, M. and Pieterse, C. M. (2008). Differential effectiveness of microbially induced resistance against herbivorous insects in Arabidopsis. Mol Plant Microbe Interact 21(7): 919-930.
- Pangesti, N., Reichelt, M., van de Mortel, J. E., Kapsomenou, E., Gershenzon, J., van Loon, J. J., Dicke, M. and Pineda, A. (2016). Jasmonic acid and ethylene signaling pathways regulate glucosinolate levels in plants during rhizobacteria-induced systemic resistance against a leaf-chewing herbivore. J Chem Ecol 42(12): 1212-1225.
- Parker, J. E. (2009). The quest for long-distance signals in plant systemic immunity. Sci Signal 2(70): pe31.
- Pieterse, C. M., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. and Bakker, P. A. (2014). Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52: 347-375.
- Pieterse, C. M., van Wees, S. C., Hoffland, E., van Pelt, J. A. and van Loon, L. C. (1996). Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8(8): 1225-1237.
- Pozo, M. J., Van Der Ent, S., Van Loon, L. C. and Pieterse, C. M. (2008). Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol 180(2): 511-523.
- Verhagen, B. W., Glazebrook, J., Zhu, T., Chang, H. S., van Loon, L. C. and Pieterse, C. M. (2004). The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol Plant Microbe Interact 17(8): 895-908.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cecchini, N. M., Song, Y., Roychoudhry, S., Greenberg, J. T. and Haney, C. H. (2019). An Improved Bioassay to Study Arabidopsis Induced Systemic Resistance (ISR) Against Bacterial Pathogens and Insect Pests. Bio-protocol 9(10): e3236. DOI: 10.21769/BioProtoc.3236.
Category
Plant Science > Plant immunity > Host-microbe interactions
Microbiology > Microbe-host interactions > Bacterium
Biochemistry > Other compound > Antimicrobial
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










