- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An Improved Staining Method for Low Signal LacZ Reporter Detection in Mouse Embryos
(*contributed equally to this work) Published: Vol 9, Iss 5, Mar 5, 2019 DOI: 10.21769/BioProtoc.3180 Views: 6709
Reviewed by: Alessandro DidonnaTharmarajan RamprasathAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Culture and Characterization of Differentiated Airway Organoids from Fetal Mouse Lung Proximal Progenitors
Zhonghui Zhang [...] Qiuling Li
Dec 5, 2024 1778 Views

Dissection and Whole-Mount Immunofluorescent Staining of Mouse Hind Paw Muscles for Neuromuscular Junction Analysis
Rebecca L. Simkin [...] James N. Sleigh
May 20, 2025 3909 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 92 Views
Abstract
In many fields of biology, especially in the field of developmental biology, LacZ reporter staining is an approach used to monitor gene expression patterns. In the LacZ reporter system, the LacZ gene is inserted in the endogenous location of the target gene via gene knock-in or by constructing a transgenic cassette in which LacZ is placed downstream of the promoter of the target gene being examined. Currently, the most common LacZ staining methods used are X-gal/FeCN staining and S-gal/TNBT staining. A serious limitation of both of these methods is that they are not effective when the LacZ gene is expressed at a low level. In an attempt to remedy this problem, we have established a new staining protocol which combines both methods. When compared to them, the method described here is better for visualizing lowly expressed genes and it has low background with high sensitivity.
Keywords: LacZ reporterBackground
The LacZ gene has been widely used as a reporter gene for detecting gene expression patterns (Stevens et al., 1989; Bonnerot and Nicolas, 1993). The protein encoded by the LacZ gene is β-galactosidase, which is able to produce visible color when incubated with specific substrates. Usually, LacZ is placed downstream of a target gene promoter in lieu of the coding sequence of the gene. Therefore, visualizing LacZ expression patterns models the endogenous expression pattern of the target gene (Figure 1A). The most popular LacZ staining approach is the X-gal/FeCN method, in which β-galactosidase catalyzes X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) hydrolysis into 5-bromo-4-chloro-3-hydroxyindole and galactose. Then, 5-bromo-4-chloro-3-hydroxyindole is oxidized into an insoluble blue dimer with the help of potassium ferricyanide and potassium ferrocyanide, which finally displays blue color (Figure 1B) (Pearson et al., 1963; Lojda, 1970). While the X-gal/FeCN staining method is highly specific and has low background, it is unable to detect genes expressed at low levels due to the low sensitivity of this stain (Gugliotta et al., 1992; Sundararajan et al., 2012). The S-gal/TNBT staining method was developed to study genes with low expression. S-gal (6-Chloro-3-indolyl-β-D-galactopyranoside), a chromogenic substrate like X-gal, can be hydrolyzed by β-galactosidase and TNBT (Tetranitro Blue Tetrazolium) to produce dark-brown formazan compounds under reducing conditions (Figure 1C). The S-gal/TNBT method is more sensitive than the X-gal/FeCN one, but a drawback is that it can have strong non-specific background staining (Sundararajan et al., 2012).
By combining both the methods described above, we developed an improved LacZ reporter staining protocol that is highly sensitive and highly specific. This method adds an additional staining step from the S-gal/TNBT method to the X-gal/FeCN method. In our staining technique, the first staining step, which is from the X-gal/FeCN method, creates an oxidative environment which consumes non-specific enzymatic activity; the second step, which is from the S-gal/TNBT method, is a reaction that is specific for β-galactosidase and results in a sensitive and specific signal. The improved method described here has been validated by detecting several genes in different embryo stages. Furthermore, this method has been used to study relatively highly expressed genes with good results: strong and specific staining, with slightly higher background than the X-gal/FeCN method (Shen et al., 2017).
Figure 1. Schematic diagram of LacZ reporters and -Galactosidase reactions. (A) The LacZ reporter gene is placed downstream of the target gene's promoter. Thus, when the expression of the target gene is induced, the gene product of LacZ, β-galactosidase, is produced. The key steps of the enzymatic reactions in the X-gal/FeCN (B) and S-gal/TNBT (C) staining methods.
Materials and Reagents
- Sterile DNase/RNase free pipette tips 10 μl, 200 μl, 1,000 μl (Labselect, catalog numbers: T-001-10, T-001-200, T-001-1000)
- 6-well plates (NUNC, catalog number: 140675)
- Embryos collected from pregnant mice at E8.5, E9.5, and E10.5 stages. The mouse strain used here has been described previously in (Shen et al., 2017)
- Paraformaldehyde (PFA) (Sigma, catalog number: P6148)
- Nonidet P-40 (NP-40) (Sangon Biotech, catalog number: A510110)
- Sodium deoxycholate (Sigma, catalog number: D6750)
- X-gal (Gold Bio, catalog number: X4281C)
- K3Fe(CN)6 (Sigma, catalog number: 244023)
- K4Fe(CN)6 (Sigma, catalog number: P3289)
- MgCl2 (Sigma, catalog number: M8266)
- EGTA (Sigma, catalog number: E3889)
- IGEPAL (Sigma, catalog number: I8896)
- Salmon-gal (S-gal) (Lab Scientific, catalog number: X668)
- Tetranitro Blue Tetrazolium (TNBT) (VWR, catalog number: TCT0250)
- NaCl (Sigma, catalog number: S7653)
- KCl (Sigma, catalog number: P9333)
- Na2HPO4 (Sigma, catalog number: S7907)
- KH2PO4 (Sigma, catalog number: P5655)
- NaH2PO4 (Sigma, catalog number: S9638)
- PBS (see Recipes)
- 4% PFA (see Recipes)
- 0.1 M phosphate buffer (pH 7.3) (see Recipes)
- Wash buffer (see Recipes)
- Staining buffer 1 (see Recipes)
- Staining buffer 2 (see Recipes)
Equipment
- Pipettes (Gilson, catalog numbers: F144801, F123600, F144058M, F123602)
- Humidified chambers (for example, covered ice boxes or foam boxes with wet paper tower)
- Dissecting forceps (Fine Science Tools, model: Dumont #5)
- Stereomicroscope (Leica, model: MZ12.5)
- Orbital Shaker (MIULAB, GS-20)
- Microbiological Incubator (Thermo Scientific, PR305225G)
- Microscope (Leica, DFC340 FX digital FireWire Camera System)
Software
- Leica Application Suite V3.7 (Image taking software for Leica DFC340 FX digital FireWire Camera System)
Procedure
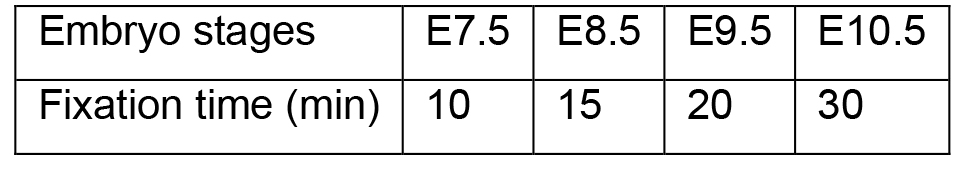
- Collect embryos from pregnant mice as reported previously (Shea and Geijsen, 2007) and fix the embryos in 10 ml 4% PFA (Recipe 2) in 6-well plates at room temperature (the volumes used here and in the following steps are for one embryo). The 6-well plates should be placed in humidified chambers, which are kept on orbital shakers set to 65 rpm. The fixation time varies and depends on the developmental stage of the embryos. The following table lists the recommended fixation times.
- Use forceps to transfer the embryos to a new well of the 6-well plate filled with 10 ml Wash Buffer (Recipe 4). Wash the embryos three times with a 10 min incubation at room temperature for each wash. During the washing steps, the 6-well plates should be placed in humidified chambers on an orbital shaker set at 65 rpm.
- Use forceps to transfer the embryos to a new well of the 6-well plate filled with 10 ml Staining Buffer 1 (Recipe 5). Incubate overnight at 37 °C in a microbiological incubator (remember to protect samples from light). The 6-well plates should be placed in humidified chambers.
- Use forceps to transfer the embryos to a new well of the 6-well plate filled with 10 ml Wash Buffer and incubate for 10 min at room temperature. Next use forceps to transfer the embryos to a new well of the 6-well plate with Staining Buffer 2 (Recipe 6) and incubate at 37 °C in a microbiological incubator. The 6-well plates should be placed in humidified chambers. The staining should be closely monitored with a stereomicroscope until specific staining appears. The staining time could vary from several minutes to one hour.
- Use forceps to transfer the embryos to a new well of the 6-well plate filled with 10 ml Wash Buffer and wash three times. Incubate each wash for 10 min. During the wash, the 6-well plates should be placed in humidified chambers, on an orbital shaker set to 65 rpm. After the embryos are washed, they can be imaged directly or kept at 4 °C in Wash Buffer for up to one week.
- Take images using Leica Application Suite V3.7 on a Leica DFC340 FX microscope. We use the Z-stack option in the Leica Application Suite V3.7 because the stained embryos are relatively thick. The images generated from the Z-stack processing are used as final results. Embryos that do not express the LacZ gene are used as negative controls. Figure 2 shows representative staining images of X-gal/FeCN, S-gal/TNBT, and the improved method described here.
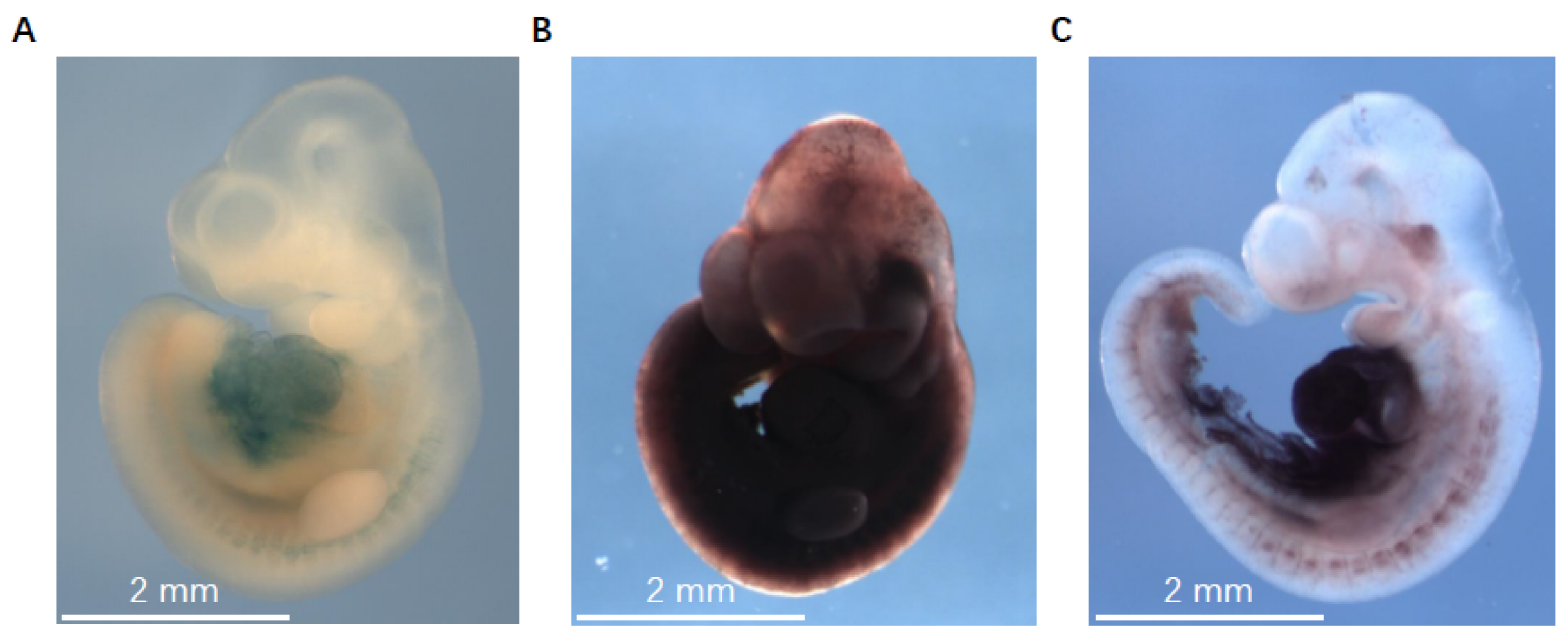
Figure 2. The representative LacZ staining images of X-gal/FeCN, S-gal/TNBT, and the improved method in detecting miR-322/-503’s expression in E10.5 embryos. A. Embryos stained with X-gal/FeCN method. B. Embryos stained with S-gal/TNBT method. C. Embryos stained with the improved method described here. The images were taken with a Leica DFC340 FX microscope at 10x magnification.
Data analysis
Scale bars, annotations, and arrows that point at the specific stained regions are added to the embryo images.
Recipes
- PBS
137 mM NaCl
2.7 mM KCl
10 mM Na2HPO4
1.8 mM KH2PO4
in distilled water, pH = 7.4 - 4% PFA
Dissolve 4% PFA in PBS
Heat at 50 °C and vortex until fully dissolved
Then adjust the pH to 7.2~7.4 - 0.1 M phosphate buffer (pH 7.3)
23% NaH2PO4 solution (0.2 M NaH2PO4 in distilled water)
77% Na2HPO4 solution (0.2 M Na2HPO4 in distilled water) - Wash buffer
0.02% NP-40
0.01% sodium deoxycholate in PBS - Staining buffer 1
5 mM K3Fe(CN)6
5 mM K4Fe(CN)6
0.02% NP-40
0.01% deoxycholate
2 mM MgCl2
5 mM EGTA
1 mg/ml X-gal in PBS - Staining buffer 2
1 mg/ml S-gal
0.4 mM TNBT
0.1% sodium deoxycholate
0.2% IGEPAL
2 mM MgCl2 in 0.1 M phosphate buffer (pH 7.3)
Acknowledgments
We thank Wei Yu at the University of Houston for guidance in animal operations; Wenjing Bao at the Liaoning University of Traditional Chinese Medicine for technical support. This work was supported by the National Natural Science Foundation of China (No. 31701289), Anhui Provincial Natural Science Foundation (No. 1808085QH234), Educational Commission of Anhui Province of China (No. KJ2017A319), Foundation for High-level Talents in Higher Education of Anhui Province of China and Start-up Funds from the Anhui Normal University.
Competing interests
All animal-related work has been approved by the Institutional Animal Care and Use Committee (IACUC).
References
- Bonnerot, C. and Nicolas, J. F. (1993). Application of LacZ gene fusions to postimplantation development. Methods Enzymol 225: 451-469.
- Gugliotta, P., Pacchioni, D. and Bussolati, G. (1992). Staining reaction for β-galactosidase in immunocytochemistry and in situ hybridization. Eur J Histochem 36(2): 143-148.
- Lojda, Z. (1970). Indigogenic methods for glycosidases. I. An improved method for β-D-glucosidase and its application to localization studies on intestinal and renal enzymes. Histochemie 22(4): 347-361.
- Pearson, B., Wolf, P. L. and Vazquez, J. (1963). A comparative study of a series of new indolyl compounds to localize β-galactosidase in tissues. Lab Invest 12: 1249-1259.
- Shea, K. and Geijsen, N. (2007). Dissection of 6.5 dpc mouse embryos. J Vis Exp (2): 160.
- Shen, X., Bao, W., Yu, W., Liang, R., Nguyen, B. and Liu, Y. (2017). An improved method with high sensitivity and low background in detecting low β-galactosidase expression in mouse embryos. PLoS One 12(5): e0176915.
- Stevens, M. E., Meneses, J. J. and Pedersen, R. A. (1989). Expression of a mouse metallothionein-Escherichia coli β-galactosidase fusion gene (MT-β gal) in early mouse embryos. Exp Cell Res 183(2): 319-325.
- Sundararajan, S., Wakamiya, M., Behringer, R. R. and Rivera-Perez, J. A. (2012). A fast and sensitive alternative for β-galactosidase detection in mouse embryos. Development 139(23): 4484-4490.
Article Information
Copyright
© 2019 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shen, X., Xu, F., Wu, S., Li, M., Zhang, J., Liang, R. and Liu, Y. (2019). An Improved Staining Method for Low Signal LacZ Reporter Detection in Mouse Embryos. Bio-protocol 9(5): e3180. DOI: 10.21769/BioProtoc.3180.
Category
Developmental Biology > Cell growth and fate > Differentiation
Stem Cell > Embryonic stem cell > Cell staining
Cell Biology > Tissue analysis > Tissue staining
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








