- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Quantification of Mouse Hematopoietic Progenitors’ Formation Using Time-lapse Microscopy and Image Analysis
Published: Vol 9, Iss 1, Jan 5, 2019 DOI: 10.21769/BioProtoc.3137 Views: 6996
Reviewed by: Wenrong HeLokesh KalekarAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Monitoring of Sperm-Independent Calcium Oscillations in Immature Oocytes of Mice
Sae Horiike [...] Hidehiko Ogawa
Feb 5, 2026 63 Views

High Content In Vitro Survival Assay of Cortical Neurons
Paolo V. Fioretti [...] Manuela Basso
Feb 5, 2026 70 Views

Simple and Rapid Model to Generate Differentiated Endometrial Floating Organoids
Adriana Bajetto [...] Tullio Florio
Feb 5, 2026 92 Views
Abstract
In vitro differentiation of mouse embryonic stem cells (mESCs) towards blood cells constitutes a well-established system to study the endothelial-to-hematopoietic transition (EHT) at the onset of blood development. Assessing the emergence of small non-adherent round blood cells in the culture without disturbing it is essential to evaluate the progression of EHT and also to test conditions potentially enhancing or repressing this process. Here, we describe how to quantify the formation of mouse hematopoietic progenitors during EHT in normal conditions or following over-expression of eight essential transcription factors using time-lapse microscopy and image analysis.
Keywords: Hematopoietic progenitorsBackground
The first hematopoietic stem and progenitor cells (HSPCs) emerge from endothelial cells in the large arteries of the mouse embryo (de Bruijn et al., 2000; Zovein et al., 2008; Chen et al., 2009;). This evolutionarily conserved event is called endothelial to hematopoietic transition (EHT). As a result of EHT, endothelial cells lose their specific markers, start to express hematopoietic genes, gain round morphology and eventually detach from the endothelial layer (Boisset et al., 2010; Kissa and Herbomel, 2010; Rybtsov et al., 2014). These specific series of events are used to detect EHT activity and monitor the progress of hematopoietic progenitor formation. The widely used method is to quantify endothelial and hematopoietic marker expressing cells. This can be done by gene expression analysis at the single-cell level (Bergiers et al., 2018). Also, protein expression change can be used to assess EHT progression by flow cytometry analysis of endothelial markers such as VE-Cadherin, CD31 and hematopoietic markers such as CD41, CD45, CD43. For spatial information, the same markers can be used for immunofluorescent staining on the fixed tissue. However, all of these techniques require harvesting of the examined cells and therefore represent a single time-point. Besides, the switch between endothelial and hematopoietic gene expressions occurs gradually and in a heterogeneous way so that a marker analysis of a specific time-point is not enough to follow the transition. To complement the gene/protein expression based detection methods, there is a need of a protocol to monitor morphological changes and quantify the round hematopoietic cell progenitors arising through EHT.
To study the hematopoietic cell formation dynamics in vitro, researchers developed embryonic stem cell differentiation protocol (Keller et al., 1993; Kennedy et al., 1997; Sroczynska et al., 2009). The protocol follows the developmental stages of hematopoiesis and recapitulates EHT in vitro (Choi et al., 1998; Nishikawa et al., 1998; Palis et al., 1999). Briefly, mouse embryonic stem cells are cultured to form embryoid bodies containing the in vitro equivalent of hemangioblast, or blast colony forming cells (BL-CFCs), which is then isolated by cell sorting. Those cells grow and give rise to smooth muscle, endothelial and hematopoietic cells (Keller et al., 1993; Faloon et al., 2000). Time-lapse imaging capturing the BL-CFC culture allows us to visualize cells transitioning into hematopoietic progenitors in real-time. Unlike the endothelial and smooth muscle cells, the cells undergoing EHT become round and bud-off from the endothelial cell layer (Lancrin et al., 2009). Here, by combining in vitro time-lapse imaging of an adherent BL-CFC culture with automatic image analysis we introduce a simple and efficient method to quantify those round cells during a culture period, which gives a direct measure of the number of cells undergoing EHT. This protocol enables us to easily test novel parameters affecting EHT rate such as over-expression of certain transcription factors (Bergiers et al., 2018) or testing pathway inhibiting small molecules in the culture media (Vargel et al., 2016). Below, we describe the details of the time-lapse microscopy of BL-CFC culture and image analysis to assess the number of cells underwent EHT.
Materials and Reagents
- Haemacytometre cover slips (Roth, catalog number: L189.1)
- BD Falcon Conical Tubes, Polypropylene, 15 ml, high-clarity, dome-seal screw cap (BD Biosciences, catalog number: 352096)
- BD Falcon Conical Tubes, Polypropylene, 50 ml, high-clarity, flat-top screw cap (BD Biosciences, catalog number: 352070)
- Costar® 6-well cell culture multiple well plate, flat bottom, with lid (Corning, catalog number: 3506)
- Millex-GP Syringe Filter Unit, 0.22 μm, polyethersulfone, 33 mm, gamma sterilized (Merck Millipore, catalog number: SLGP033RS)
- Mμlti®-safety microcentrifuge tubes, SafeSeal® Tubes (Carl Roth, catalog number: 7080.1)
- Stericup GP 0.2 μm/150 ml (Merck-Millipore, catalog number: SCGPU01RE)
- Stericup GP 0.2 μm/500 ml (Merck-Millipore, catalog number: SCGPU05RE)
- TipOne® 10 μl Graduated Filter Tip (Sterile), Refill (Starlab, catalog number: S1121-2710)
- TipOne® 1,000 μl Graduated Filter Tip (Sterile), Refill (Starlab, catalog number: S1122-1730)
- TipOne® 20 μl Graduated Filter Tip (Sterile), Refill (Starlab, catalog number: S1120-1710)
- TipOne® 200 μl Graduated Filter Tip (Sterile), Refill (Starlab, catalog number: S1120-8710)
- i8TFs mESC line (Bergiers et al., 2018)
- D4T endothelial cells (Choi et al., 1998)
- Ascorbic acid (Sigma, catalog number: A4544)
- Bovine Serum Albumin (BSA) (Sigma, catalog number: A9418)
- Doxycycline (Sigma, catalog number: D9891)
- Fetal bovine serum (FBS) (PAA, catalog number: A15-102)
- Gelatin (BDH, catalog number: 440454B)
- IL-6 (R and D, catalog number: 406 ML)
- IMDM (Lonza, catalog number: BE12-726F)
- L-glutamine (Gibco, catalog number: 25030-024)
- Monothioglycerol (MTG) (Sigma, catalog number: M6145)
- OxoidTM Phosphate Buffered Saline (PBS) Tablets (Thermo Scientific, catalog number: BR0014G)
- Transferrin (Roche, catalog number: 10652202001)
- VEGF (R and D, catalog number: 293-VE)
- Distilled Water (Thermo Scientific, GibcoTM, catalog number: 15230188)
- 0.1% gelatin solution (see Recipes)
- 10 mg/ml doxycycline stock solution (see Recipes)
- 5 mg/ml ascorbic acid stock solution (see Recipes)
- D4T endothelial cell supernatant (see Recipes)
- PBS + 0.1% BSA solution (see Recipes)
- 10 μg/ml VEGF stock solution (see Recipes)
- 10 μg/ml IL6 stock solution (see Recipes)
- Conditioned IMDM (see Recipes)
- IMDM + 20% FBS (see Recipes)
- MTG dilution (see Recipes)
- BL-CFC culture medium (see Recipes)
Equipment
- Falcon® 10 ml Serological Pipet, Polystyrene, 0.1 Increments, Individually Packed, Sterile (Corning, catalog number: 357551)
- Falcon® 2 ml Aspirating Pipet, Polystyrene, without Graduations, Individually Wrapped, Sterile (Corning, catalog number: 357558)
- Falcon® 5 ml Serological Pipet, Polystyrene, 0.1 Increments, Individually Packed, Sterile (Corning, catalog number: 357543)
- 37 °C, 5% CO2 cell culture incubator (Thermo Scientific, model: Series II Water Jacket)
- Biosafety cabinet (Tissue culture hood) (Thermo Scientific, model: MSC Advantage)
- Brightfield inverted Microscope (Leica, model: DMIL LED Inverted)
- Centrifuge (Eppendorf, model: 5810 R)
- Computer (Dell Precision T3500)
- IncuCyte HD (Essen Biosciences)
- Neubauer counting chamber improved (Roth, catalog number: T729.1)
- Pipetman P10, 1 to 10 μl (Gilson, catalog number: F144802)
- Pipetman Starter Kit (P20, P200, P1000) (Gilson, catalog number: F167300)
- Pipette controller, PIPETBOY acu 2 (VWR, catalog number: 612-0928)
- Standard -20 °C freezer (LIEBHERR, model: LCv 4010 MediLine)
- Standard -80 °C freezer (Heraeus, model: HFU586 Top Freeze)
- Standard fridge (LIEBHERR, model: LKUexv 1610 MediLine)
- Vacuum pump BVC control (Vacuubrand, catalog number: 20727200)
Software
- CellProfiler (Kamentsky et al., 2011; www.cellprofiler.org)
- Fiji (Schindelin et al., 2012; http://fiji.sc/Fiji)
- IncuCyte software (Essen Biosciences, 2011A Rev2)
Procedure
Prior to the start of the BL-CFC culture, ESCs are differentiated according to previously described protocols (Sroczynska et al., 2009). Flk1+ cells are isolated using magnetic sorting following the manufacturer’s instructions (Miltenyi Biotech). The procedure described below has been optimized for adherent BL-CFC culture allowing the use of time-lapse microscopy. The quantities and volumes listed were further adapted for the comparison of two culture conditions: with or without doxycycline.
Note: Before starting, prepare working solutions as described in “Recipes”. Careful, BL-CFC culture medium should be prepared fresh.
Day 0
- Put 1 ml/well of 0.1% gelatin solution (see Recipes) into eight wells of Costar® 6-well cell culture plates (one plate of four wells for each line) and leave at room temperature (RT) for at least 20 min.
- For each line, introduce 0.425 million Flk1+ cells obtained after embryoid body differentiation of ESCs and counted using a Neubauer counting chamber (Sroczynska et al., 2009) into a 15-ml Falcon tube containing 5 ml of IMDM + 20% FBS.
- Centrifuge at 290 x g for 5 min.
- For each line, remove the supernatant using an aspirating pipet connected to the vacuum pump BVC control, gently tap the bottom of the tube to loosen the cell pellet and resuspend into 8.5 ml of BL-CFC culture medium.
Note: Always use individual pipets and tips for each cell line to avoid cross-contamination. - Aspirate the gelatin from the pre-coated Costar® 6-well cell culture plate wells using an aspirating pipet.
- Resuspend and evenly distribute 2 ml/well of cell suspension into four gelatin-coated wells for each cell lines.
Note: This means that 0.1 million Flk1+ cells are added per well. - Agitate the plates parallel to the bench doing vertical and horizontal small jolting movements.
Note: Careful not to do circular movements which would result in the formation of a vortex and, consequently, in all the cells going to the center of the well. - Place the plate into a 37 °C, 5% CO2 cell culture incubator for 24 h.
- Keep the BL-CFC culture medium in the fridge for the doxycycline dilution on the next day.
Note: The differentiation rate can be affected by cell confluence. Make sure the cells are equally spread across the surface of the wells to insure that the emergence of round blood cells will occur at the same rate everywhere in the wells.
Day 1
- Prepare 0.2 μg/μl doxycycline dilution by adding 4 μl of 10 mg/ml doxycycline stock solution (see Recipes) to 196 μl of BL-CFC culture medium kept in the fridge from the day before. Mix well.
- Take out the culture plates from the incubator and check under the microscope that the wells are all similar to each other.
- Add 10 μl of 0.2 μg/μl doxycycline dilution to each +dox condition well (two wells for each line at a final doxycycline concentration of 1 μg/ml). Add the doxycycline directly in the center of the wells and mix directly after by agitating the culture plates carefully.
- Add 10 μl of the BL-CFC culture medium to each -dox condition well as control (two wells for each line).
- Put back the two plates inside the incubator and place them carefully into the IncuCyte HD microscope.
Note: Don’t write on top of the wells and avoid splashes of the medium on the lid to prevent malfunctioning of the IncuCyte HD as it takes pictures from the top. - While the lid condensation is going away, set up the IncuCyte HD device using the IncuCyte software on the computer:
- Connect to the device and log in.
- Click on “Schedule Upcoming Scans” in the Task List panel on the left (see Figure 1 A).
- In the Drawer Setup window, select an empty vessel on the screen, right-click on the location where you inserted your first plate and click on “New” (see Figure 1).
- Keep this vessel selected and select the following settings in the Scan Setup tab, on the left side of the drawer map (see Figure 1 B):
Tray Type: Microplates
Vessel Type: 6-well Corning
Scan Type: Phase Contrast
Scan Pattern: Select the appropriate scan pattern or create one clicking on “Edit Scan Patterns” on the bottom left of the window (see Figure 1 C). Select 16 images/well for each 6-well.
Note: The IncuCyte can take up to 121 images per well in a 6-well plate format. However, by choosing this option, we would have increased the time of imaging considerably, therefore leading to time points spaced by at least two hours instead of the 15 min needed to capture the morphological changes occurring during the BL-CFC culture. - With the vessel selected, go to the Properties tab (see Figure 1 D) and fill the following fields (see Figure 2 A):
Label: Name of your experiment/vessel type
Cell type: Exact name of the cell line/cell type used
Passage: Number of passages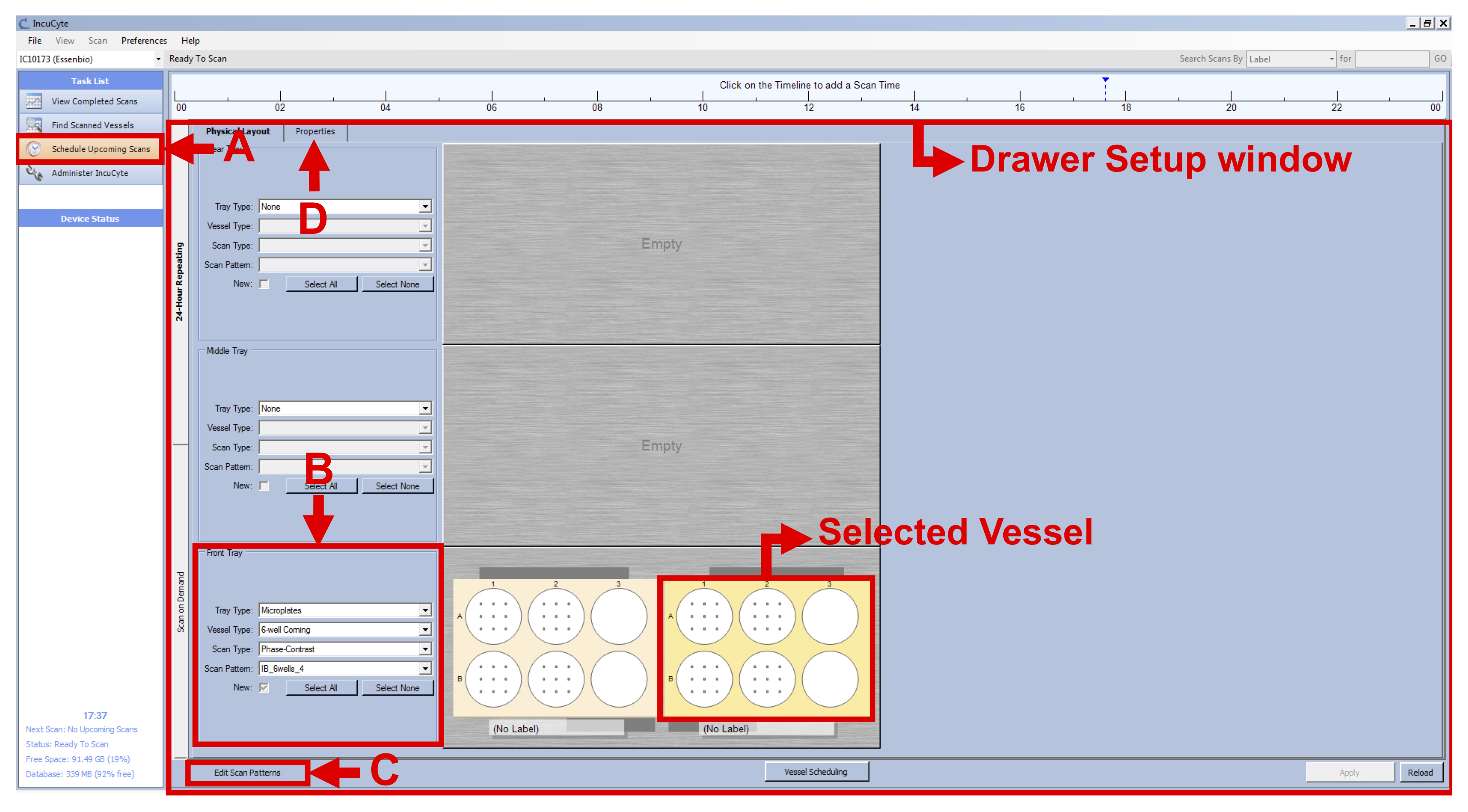
Figure 1. IncuCyte setup - Click on “Plate Map” (see Figure 2 B) and in the new window set up the plate map of your experiment (see Figure 3):
- Add a new compound and name it “+dox” (see Figure 3 A).
- Select the +dox compound, the wells that you treated with doxycycline and click on “Add +dox” (see Figure 4 A).
- Select the concentration and click on OK.
- Click on OK.
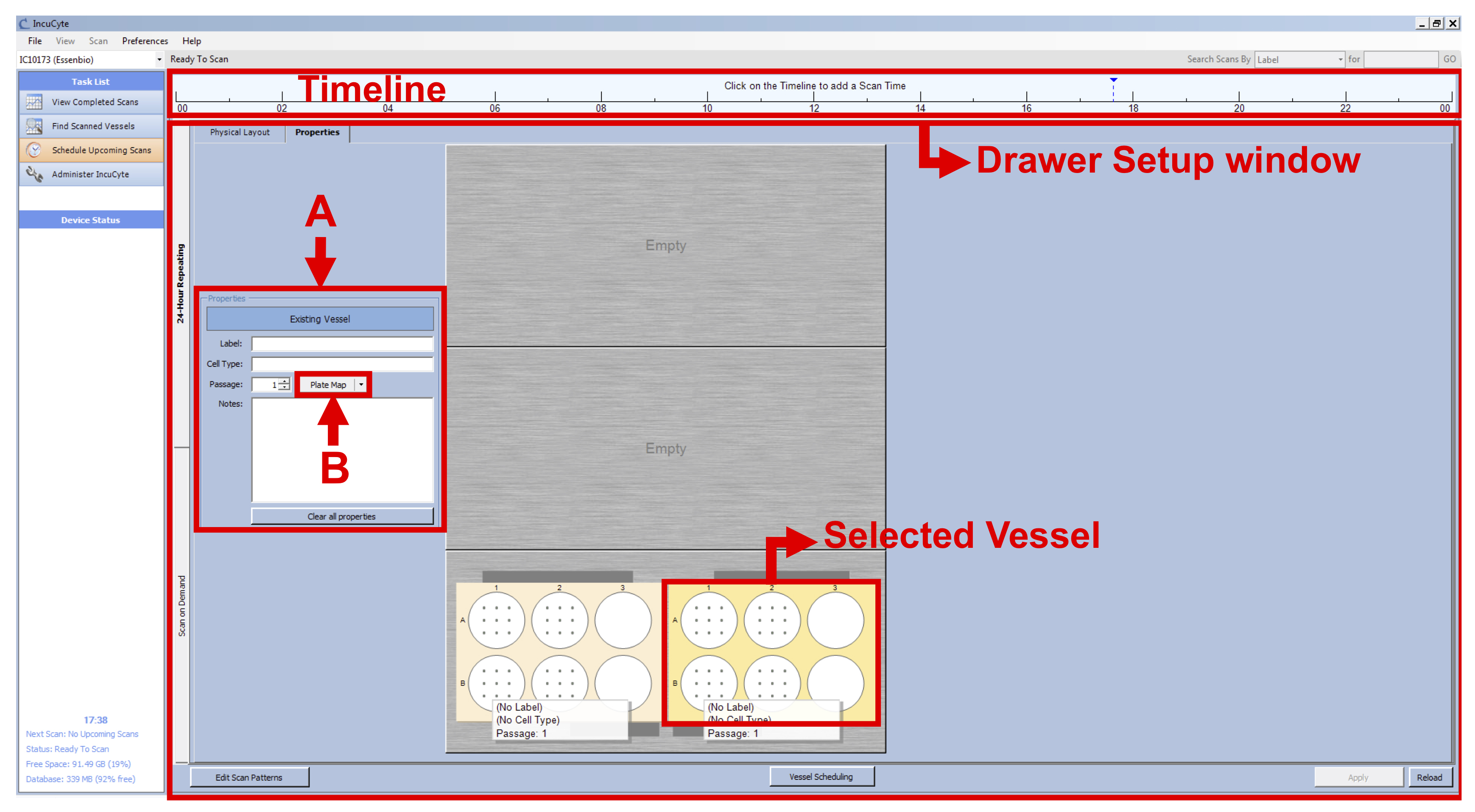
Figure 2. IncuCyte Properties setup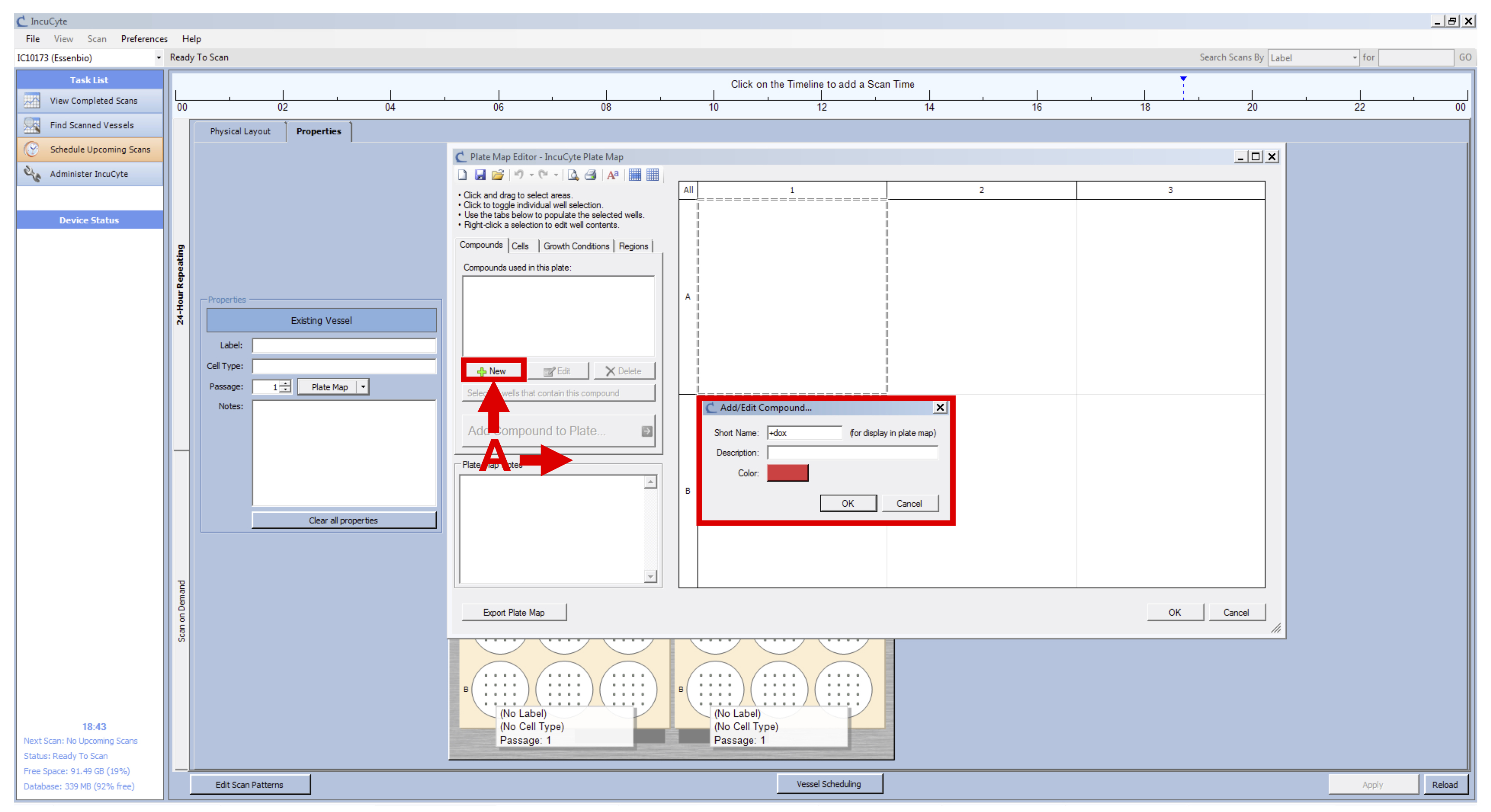
Figure 3. IncuCyte Plate Map initial setup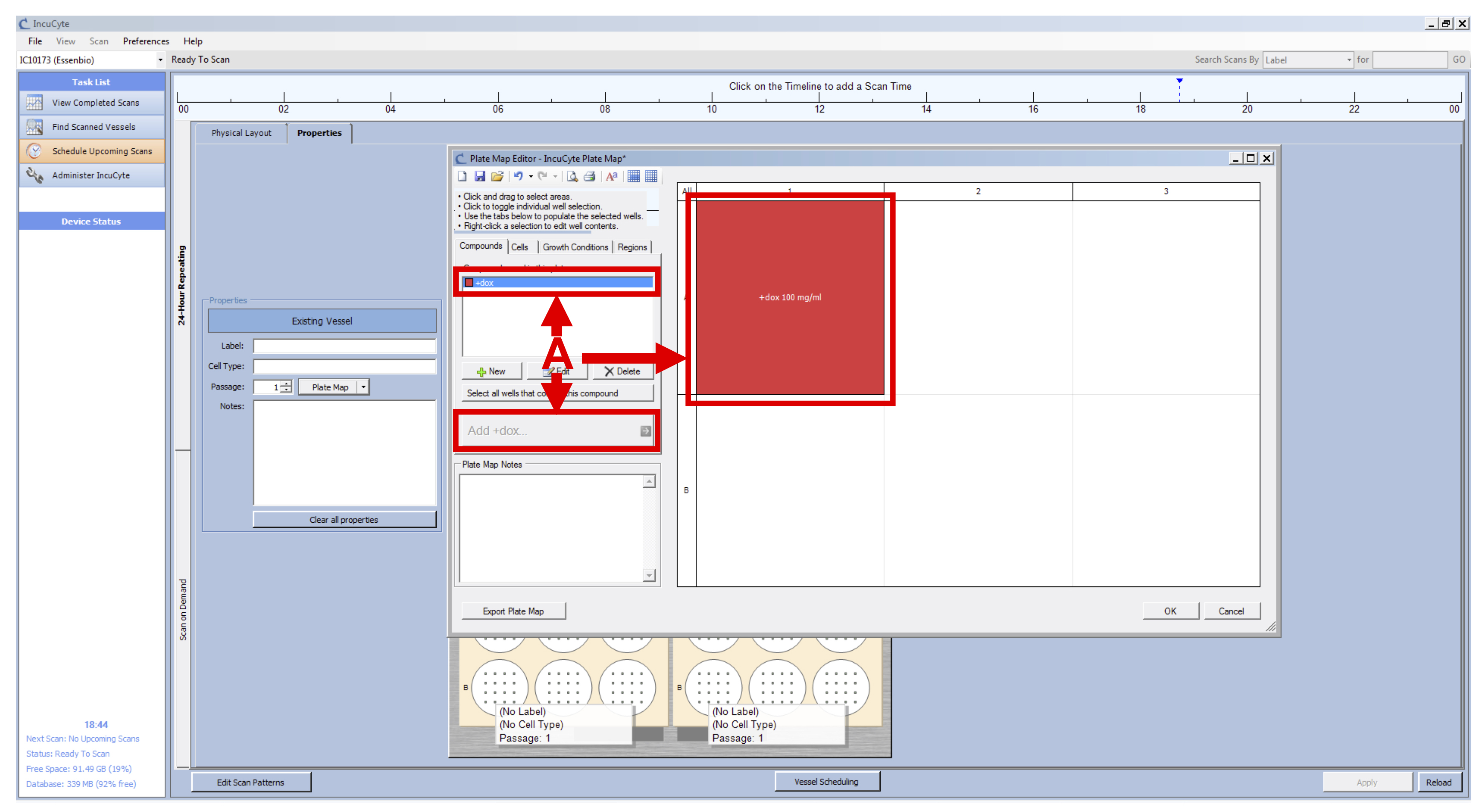
Figure 4. IncuCyte Plate Map setup - Repeat the setup of the vessel (from Day1 Steps 6c-6f) for your second plate.
- Right-click on the upper timeline (see Figure 2) and click on “Set intervals”.
- Set intervals every 15 min starting from time 0, for a total of 24 h.
Note: Even with a total of 24 h, the Incucyte device will scan your vessels every 15 min until you manually remove your vessels from the software. - Check the grey bars to make sure they are not overlapping (see Figure 5 A), check that IncuCyte device is not scanning (see Figure 5 B) and click on “Apply” (see Figure 5 C).
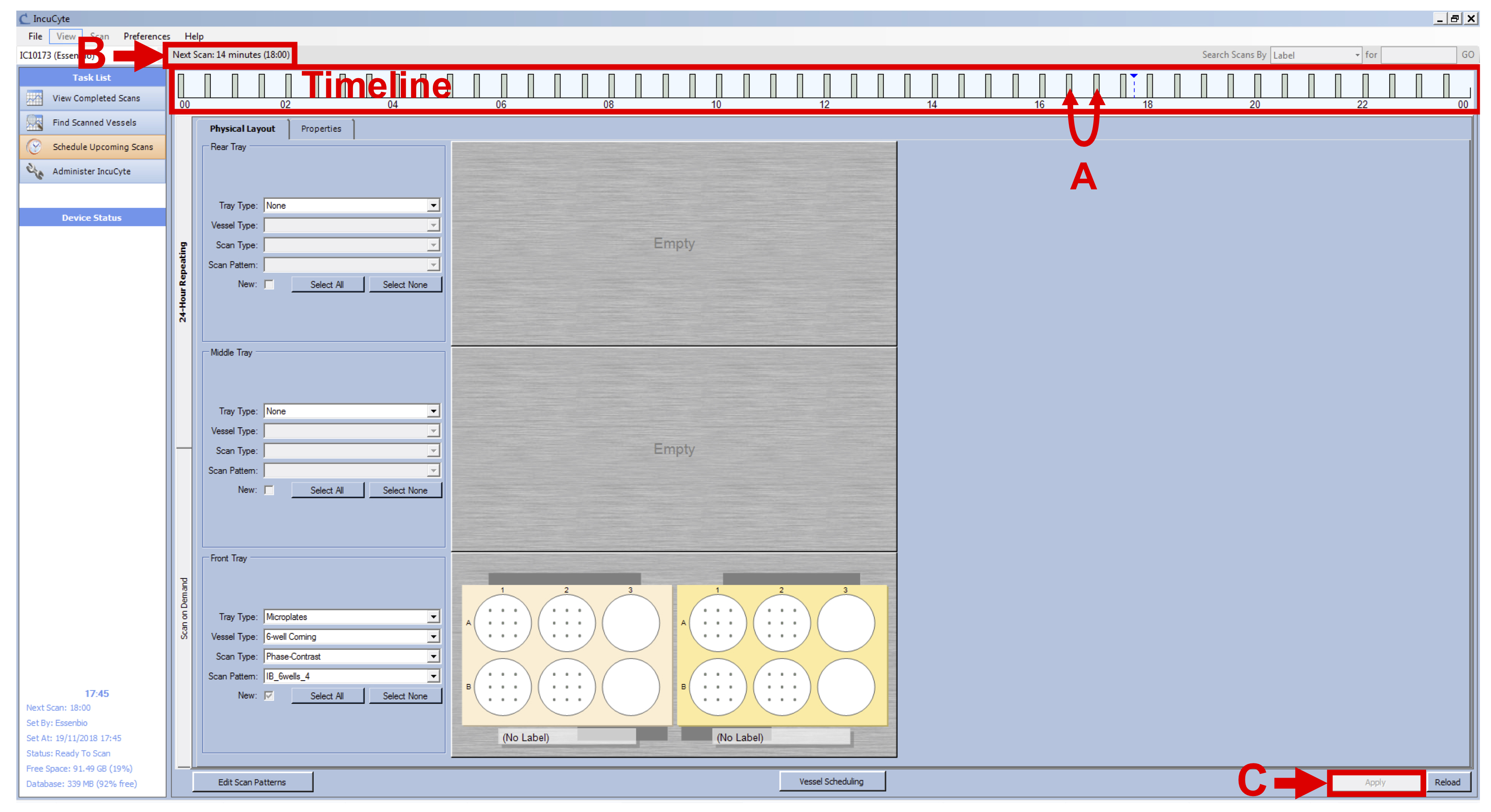
Figure 5. Set Intervals
- Come back after 15 min to check that everything is working and switch off the computer if you wish.
Day 3
- After 48-h treatment, click on “Schedule Upcoming Scans” in the Task List panel of the IncuCyte software.
- Right-click on the upper timeline and select “Delete Intervals”.
- Click on “Apply” to stop the scanning by IncuCyte.
- Take out your plates and proceed with flow cytometry analysis according to standard protocols.
Data analysis
- Export your images from the IncuCyte software:
- Click on “Find Scanned Vessels” in the Task List panel of the IncuCyte software (see Figure 6 B).
- Select one of your cell line vessels (see Figure 6 C) and click on “View Vessel” button in the lower right-hand corner of the screen (see Figure 6 D).
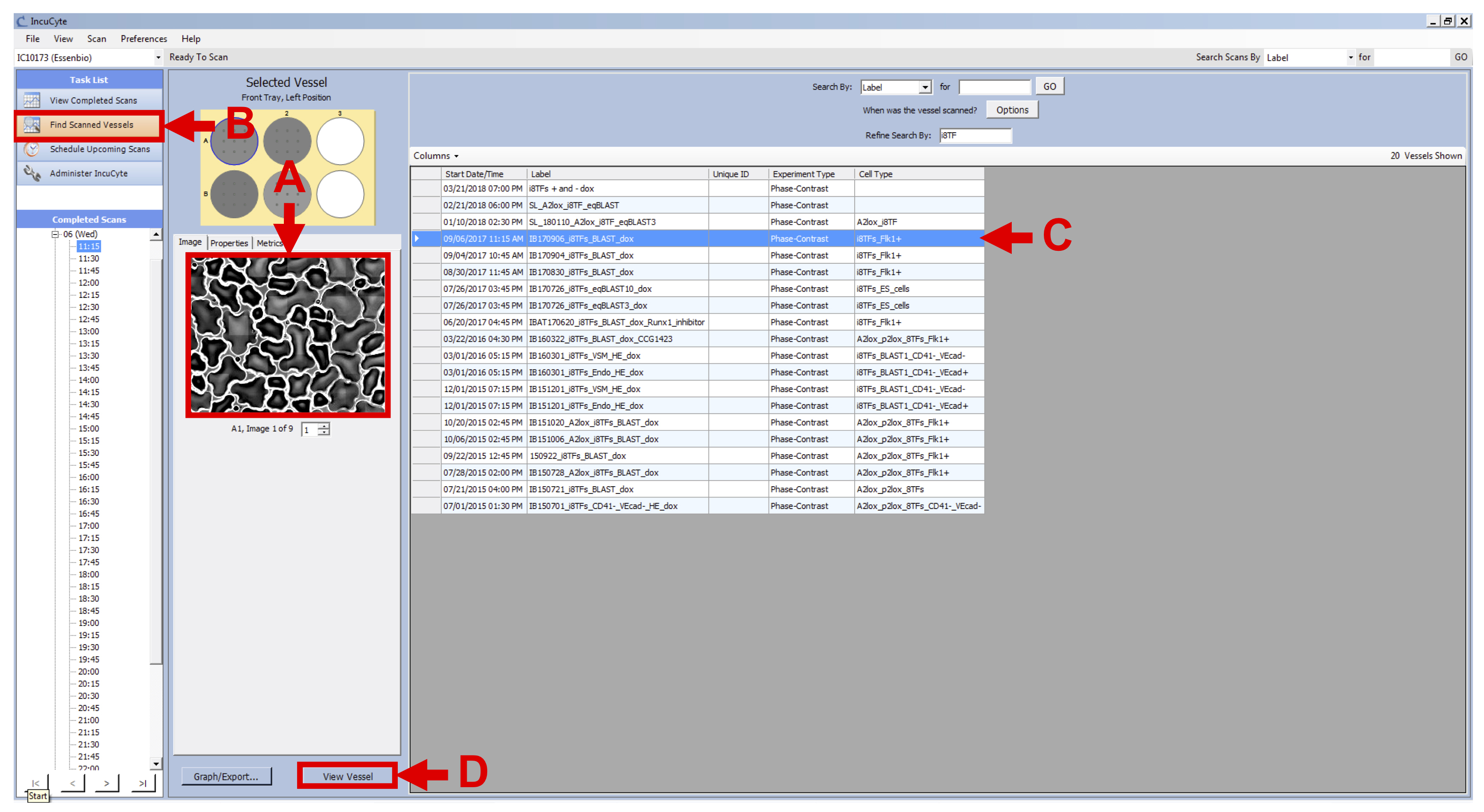
Figure 6. View Vessels - Check that the scans are fine (no dust, condensation, etc.).
- Selecting “Export Movie or Image Set…” from the “Utilities” pull down menu (see Figure 7 A).
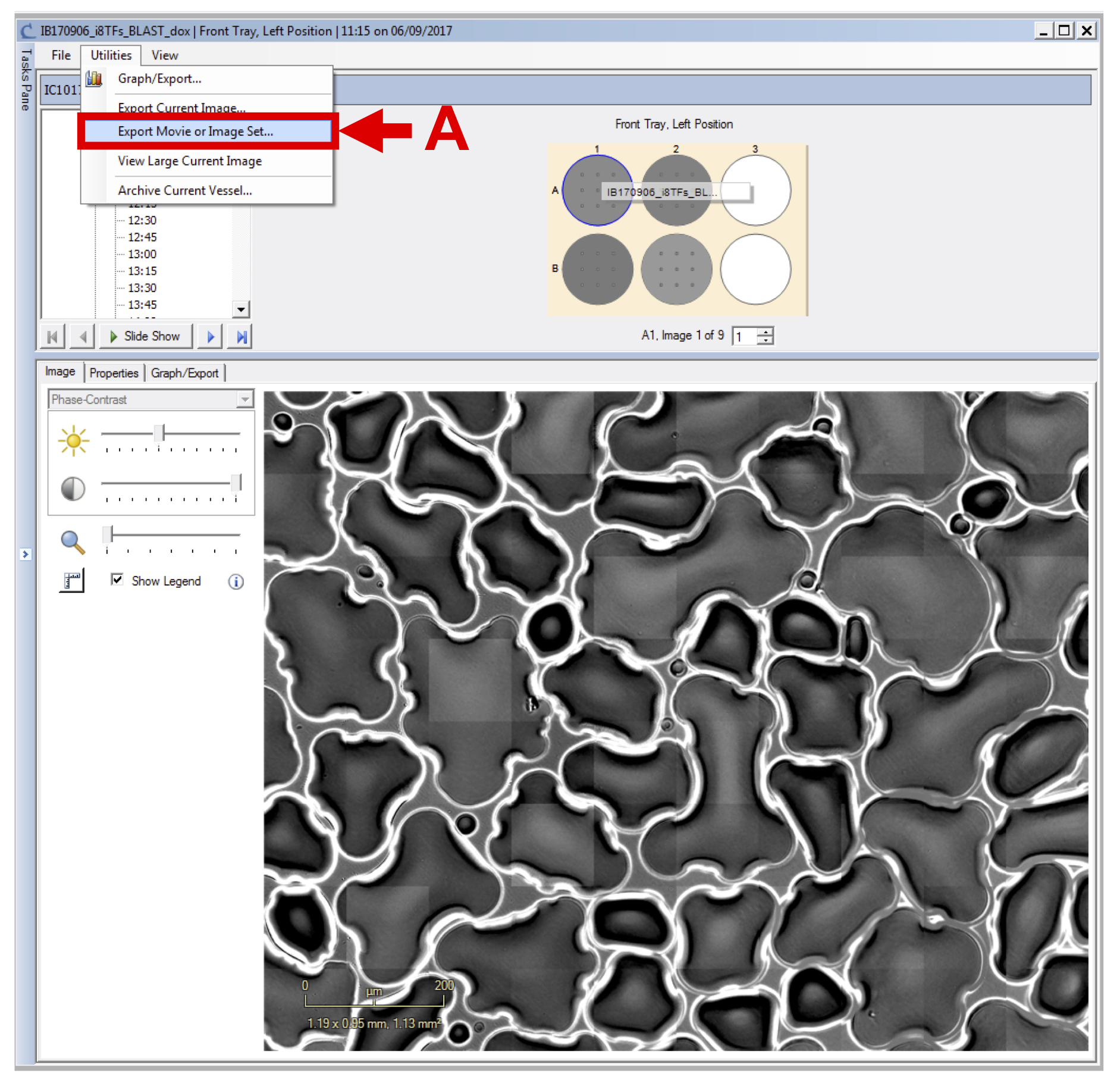
Figure 7. Export image set - In the new window, select the appropriate time frame (see Figure 8 A), the wells corresponding to one condition (see Figure 8 B), all images (see Figure 8 C), select the sequence type labeled “set of individual images” (see Figure 8 D) and export phase-contrast original image (.JPEG).
- Specify the destination folder and the prefix of the files that will be generated (see Figure 8 E).
Note: Create one folder per condition per cell line. - Click on “Export … files” (see Figure 8 F).
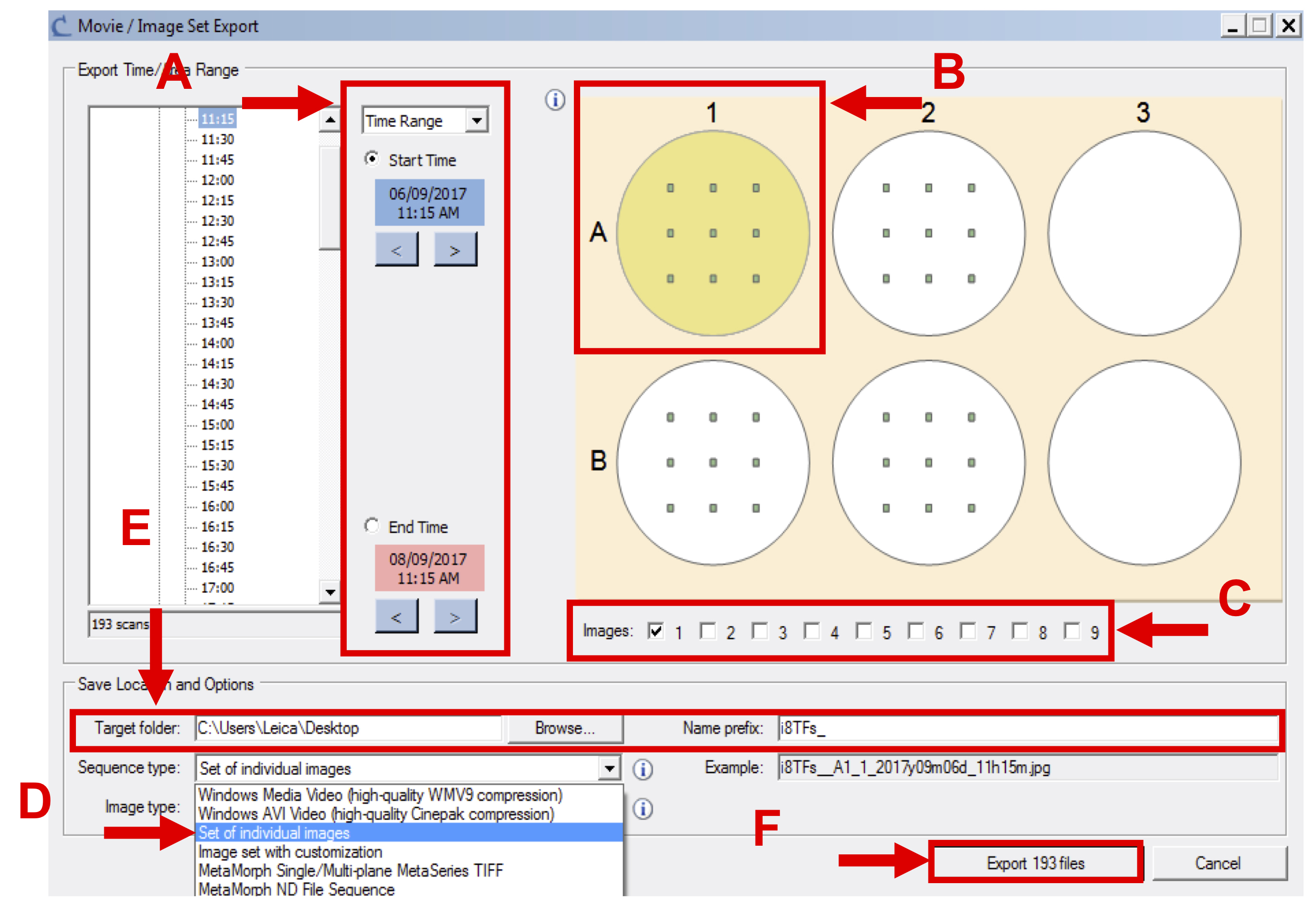
Figure 8. Final step of export image set - Repeat Steps 1a-1h for all the vessels and conditions to be analyzed (see Supplemental File 1: Example dataset of IncuCyte images for the i8TF cell line for an example dataset).
- Analyze your images using CellProfiler software to get the number of round cells for each time-point, cell line and condition:
- Install CellProfiler2.2.0, which can be downloaded from here: http://cellprofiler.org/previous_releases/
Note: You need Java as well: http://cellprofiler.org/releases/. - Open CellProfiler and load the CellProfiler pipeline (see Supplemental File 2: CellProfiler pipeline for round cell counting) using File > Import > Pipeline from File… in the main menu of CellProfiler.
- Drag and drop the whole IncuCyte export destination folder into the File list area of the Images module (see Figure 9 A).
- Click on “Analyze images” on the bottom left to start processing (see Figure 9 B).
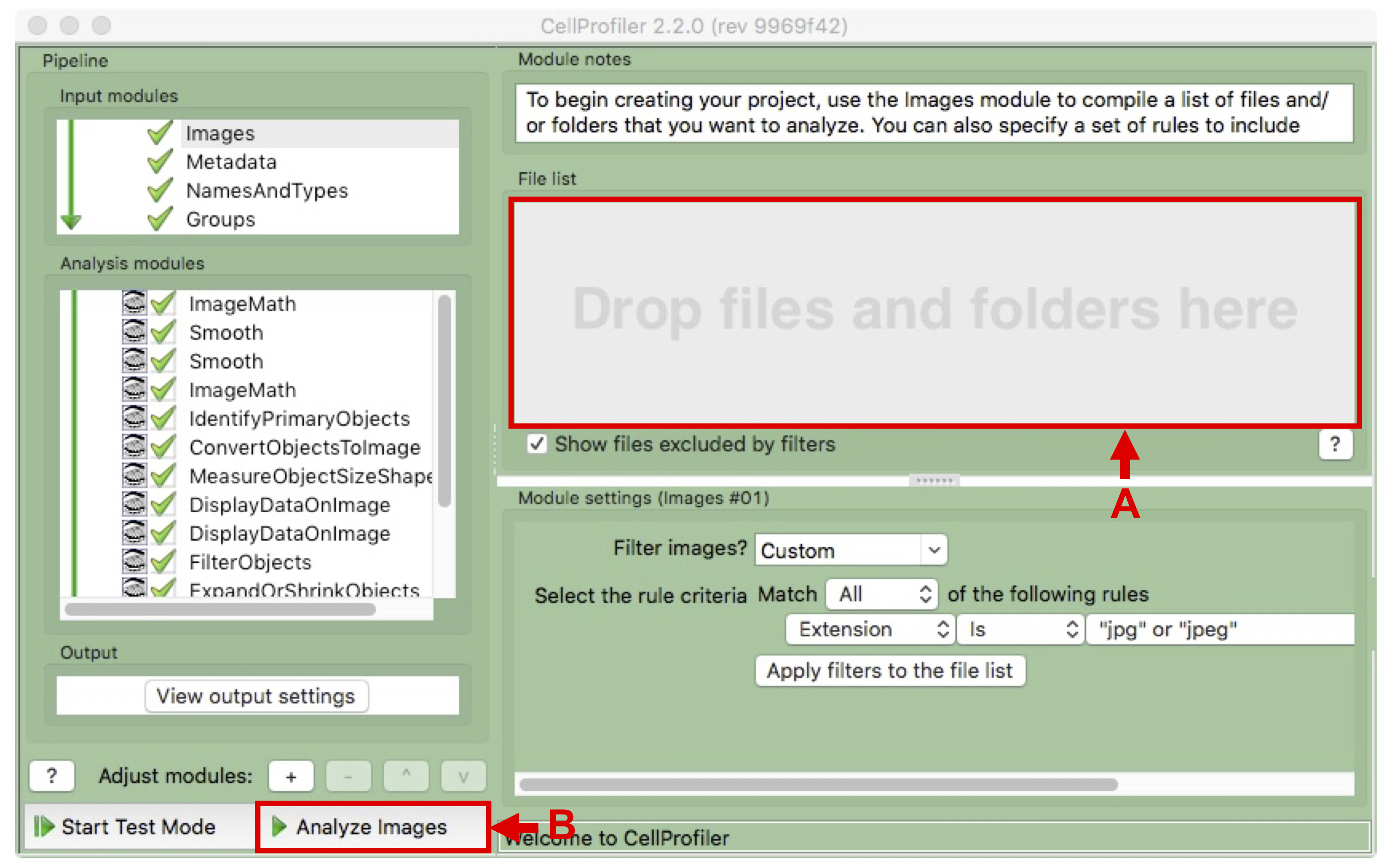
Figure 9. Image module of CellProfiler- Use the Browser to access the output “image.txt” file in the output folder (as an example, see Supplemental File 3: CellProfiler output files for the example dataset) and open it with Excel or in R to visualize the results as graphs. In the output .txt file, the Count_RoundCells column contains the round cell numbers. The ImageNumber column contains the order in which the images were analyzed by CellProfiler. The Metadata_condition column contains the prefix given while exporting the images from the IncuCyte software. The Metadata_day column contains the day in which the images were taken. The Metadata_filename column contains the file names of the images analyzed such as exported by the IncuCyte software. The Metadata_foldername column corresponds to the input folder. The Metadata_position column contains the position of the images inside the well. The Metadata_time column contains the time in which the images were taken. The Metadata_well column contains the name of the well from which the images were taken. Each row corresponds to one image.
- See Figures 10 and 11, and Supplemental File 3: CellProfiler output files for the example dataset for the analysis output of the example dataset.
Notes:- The Metadata extraction described above relies on the IncuCyte file naming scheme. If this naming scheme is changing, e.g., due to version updates by IncuCyte, or because you are using a different microscope, it will not work. In such cases please contact us and we will help you adapting the Metadata extraction inside CellProfiler.
- In the output folder, CellProfiler also generates copies of the individual images with all counted round cells marked by a yellow dot (Figure 10).
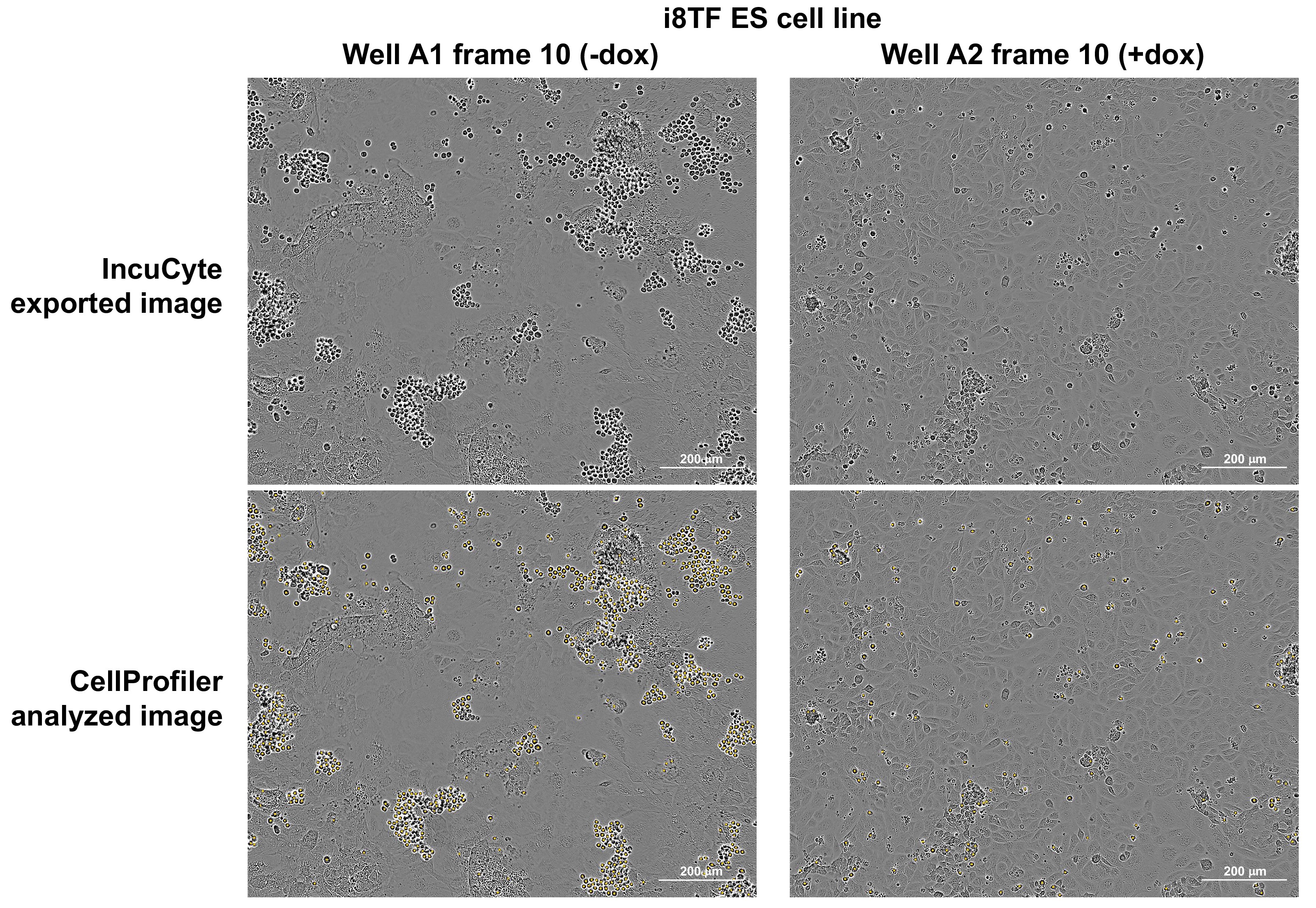
Figure 10. IncuCyte images before and after CellProfiler analysis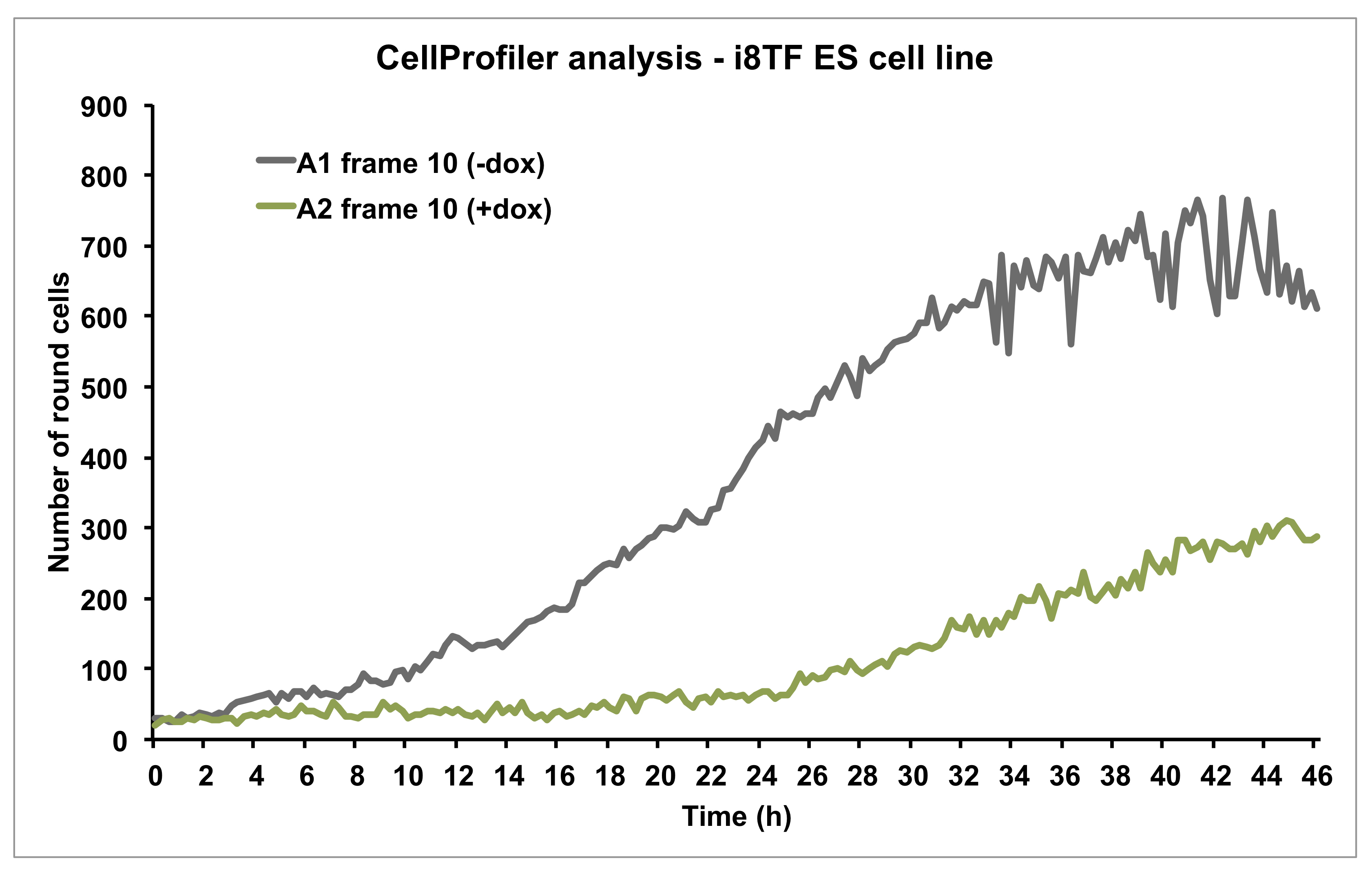
Figure 11. Graph showing the number of round cells over time as calculated by CellProfiler
- Install CellProfiler2.2.0, which can be downloaded from here: http://cellprofiler.org/previous_releases/
- Generate movies from your images using Fiji software to illustrate differences between round cell emergence rates and highlight other morphological variations:
- Install Fiji software, which can be downloaded from here: http://fiji.sc/Fiji.
- Open Fiji.
- In your Browser, sort the IncuCyte exported files by Name, select all the files corresponding to the images taken from one frame of one condition for one cell line. By clicking on the last image, drag and drop all the files in Fiji. Let Fiji open the files without changing the order of the opened windows.
- To create a stack, select Image > Stacks > Images to Stack and then click on OK in the new window.
- Check the proper alignment of the stack slices by clicking on the play icon on the bottom left of the image. If the movie is readable, go directly to Step 3h.
- To align the slices, select Plugins > Image Stabilizer. In the new window, insert the following values:
Transformation: Translation
Maximum Pyramid Levels: 1
Template Update Coefficient (0-1): 0.90
Maximum Iterations: 200
Error Tolerance: 0.0000001
Select Output to a New Stack and click on OK. - Check the slice alignment of the stabilized stack by clicking on the play icon on the bottom left of the image. If the movie is readable, go to Step 3h otherwise repeat Step 3f until all the slices are properly aligned (up to 3-4 times).
- Save the movie by selecting File > Save As > AVI…. In the new window, select JPEG Compression and 10 fps as Frame Rate.
- See Videos 1 and 2 based on the example dataset.Video 1. BL-CFC culture in normal conditions. The video is a 48-h-time-lapse microscopy analysis of BL-CFC culture in absence of doxycycline.Video 2. BL-CFC culture following over-expression of eight key transcription factors. The video is a 48-h-time-lapse microscopy analysis of BL-CFC culture in presence of doxycycline, i.e., following the over-expression of eight key transcription factors (Runx1, Cbfb, Gata2, Tal1, Fli1, Lyl1, Erg and Lmo2).
Recipes
Note: Prepare solutions 1-9 in advance.
- 0.1% gelatin solution (stored at 4 °C)
0.2 g of gelatin
200 ml of PBS
Note: Dissolve the powder, sterile filter with Stericup GP 0.2 μm/500 ml, aliquot and store at 4 °C. - 10 mg/ml doxycycline stock solution (stored at -20 °C)
10 mg of doxycycline
1 ml of sterile distilled water
Note: Dissolve the powder, aliquot and store at -20 °C. Always freshly thaw an aliquot. - 5 mg/ml ascorbic acid stock solution (stored at -20 °C)
0.5 g of ascorbic acid
100 ml of distilled and sterile water
Note: Dissolve the powder, sterile filter with a Stericup GP 0.2 μm/150ml, aliquot and store at -20 °C. Always freshly thaw an aliquot. - D4T endothelial cell supernatant (stored at -20 °C)
Prepare D4T endothelial cell supernatant following the procedure described by Choi and colleagues (Choi et al., 1998).
Note: Prepare, aliquot and store at -20 °C. Once thawed, can be stored at 4 °C and used up to one month after. - PBS + 0.1% BSA solution
0.05 g of BSA
50 ml of PBS
Note: Dissolve, sterile filter using Millex-GP Syringe Filter, aliquot and store at -20 °C. - 10 μg/ml VEGF stock solution (stored at -80 °C)
Dissolve a 10 μg vial in 1 ml of sterile PBS + 0.1% BSA solution
Note: Dissolve, aliquot and store at -80 °C. Once thawed, can be stored at 4 °C and used up to one month after. - 10 μg/ml IL6 stock solution (stored at -80 °C)
Dissolve a 10 μg vial in 1 ml of sterile PBS + 0.1% BSA solution
Note: Dissolve, aliquot and store at -80 °C. Once thawed, can be stored at 4 °C and used up to one month after. - Conditioned IMDM (stored at 4 °C)
1 bottle of IMDM
5 ml of L-glutamine
5 ml of Penicillin-streptomycin - IMDM + 20% FBS (stored at 4 °C)
30 ml of FBS
120 ml of conditioned IMDM
Note: Sterile filter using StericupTM 150 ml bottle. - MTG dilution (to be prepared fresh)
13 μl of MTG
1 ml of conditioned IMDM
Note: Careful, MTG is viscous. Mix well after dilution. - BL-CFC culture medium (to be prepared fresh)
14.49 ml of conditioned IMDM
2 ml of FBS
0.2 ml of L-glutamine
0.12 ml of Transferrin
0.06 ml of MTG dilution
0.1 ml of 5 mg/ml acid ascorbic stock solution
3 ml of D4T endothelial cell supernatant
0.01 ml of 10 μg/ml VEGF stock solution
0.02 ml of 10 μg/ml IL6 stock solution
Note: Prepare fresh in a 50 ml Falcon tube and sterile filter using Millex-GP Syringe Filter.
Acknowledgments
The EMBL Interdisciplinary Postdocs (EIPOD) Initiative (Post-doc fellowship) funded Isabelle Bergiers. The European Molecular Biology Laboratory has funded this work.
Competing interests
The authors do not have any conflicts of interests or competing interests.
References
- Bergiers, I., Andrews, T., Vargel Bolukbasi, O., Buness, A., Janosz, E., Lopez-Anguita, N., Ganter, K., Kosim, K., Celen, C., Itir Percin, G., Collier, P., Baying, B., Benes, V., Hemberg, M. and Lancrin, C. (2018). Single-cell transcriptomics reveals a new dynamical function of transcription factors during embryonic hematopoiesis. Elife 7: e29312.
- Boisset, J. C., van Cappellen, W., Andrieu-Soler, C., Galjart, N., Dzierzak, E. and Robin, C. (2010). In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464(7285): 116-120.
- Chen, M. J., Yokomizo, T., Zeigler, B. M., Dzierzak, E. and Speck, N. A. (2009). Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457(7231): 887-891.
- Choi, K., Kennedy, M., Kazarov, A., Papadimitriou, J. C. and Keller, G. (1998). A common precursor for hematopoietic and endothelial cells. Development 125(4): 725-732.
- de Bruijn, M. F., Speck, N. A., Peeters, M. C. and Dzierzak, E. (2000). Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J 19(11): 2465-2474.
- Faloon, P., Arentson, E., Kazarov, A., Deng, C. X., Porcher, C., Orkin, S. and Choi, K. (2000). Basic fibroblast growth factor positively regulates hematopoietic development. Development 127(9): 1931-1941.
- Kamentsky, L., Jones, T.R., Fraser, A., Bray, M., Logan, D., Madden, K., Ljosa, V., Rueden, C., Harris, G.B., Eliceiri, K., Carpenter, A.E. (2011). Improved structure, function, and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27(8):1179-1180.
- Keller, G., Kennedy, M., Papayannopoulou, T. and Wiles, M. V. (1993). Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol 13(1): 473-486.
- Kennedy, M., Firpo, M., Choi, K., Wall, C., Robertson, S., Kabrun, N. and Keller, G. (1997). A common precursor for primitive erythropoiesis and definitive haematopoiesis. Nature 386(6624): 488-493.
- Kissa, K. and Herbomel, P. (2010). Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464(7285): 112-115.
- Lancrin, C., Sroczynska, P., Stephenson, C., Allen, T., Kouskoff, V. and Lacaud, G. (2009). The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature 457(7231): 892-895.
- Nishikawa, S. I., Nishikawa, S., Hirashima, M., Matsuyoshi, N. and Kodama, H. (1998). Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hemopoietic lineages. Development 125(9): 1747-1757.
- Palis, J., Robertson, S., Kennedy, M., Wall, C. and Keller, G. (1999). Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126(22): 5073-5084.
- Rybtsov, S., Batsivari, A., Bilotkach, K., Paruzina, D., Senserrich, J., Nerushev, O. and Medvinsky, A. (2014). Tracing the origin of the HSC hierarchy reveals an SCF-dependent, IL-3-independent CD43- embryonic precursor. Stem Cell Reports 3(3): 489-501.
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., Preibisch, S., Rueden, C., Saalfeld, S., Schmid, B., Tinevez, J.Y., White, D.J., Hartenstein, V., Eliceiri, K., Tomancak, P., Cardona, A. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676-682.
- Sroczynska, P., Lancrin, C., Pearson, S., Kouskoff, V. and Lacaud, G. (2009). In vitro differentiation of mouse embryonic stem cells as a model of early hematopoietic development. Methods Mol Biol 538: 317-334.
- Vargel, O., Zhang, Y., Kosim, K., Ganter, K., Foehr, S., Mardenborough, Y., Shvartsman, M., Enright, A. J., Krijgsveld, J. and Lancrin, C. (2016). Activation of the TGFβ pathway impairs endothelial to haematopoietic transition. Sci Rep 6: 21518.
- Zovein, A. C., Hofmann, J. J., Lynch, M., French, W. J., Turlo, K. A., Yang, Y., Becker, M. S., Zanetta, L., Dejana, E., Gasson, J. C., Tallquist, M. D. and Iruela-Arispe, M. L. (2008). Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 3(6): 625-636.
Article Information
Copyright
Bergiers et al. This article is distributed under the terms of the Creative Commons Attribution License (CC BY 4.0).
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Bergiers, I., Tischer, C., Bölükbaşı, �. V. and Lancrin, C. (2019). Quantification of Mouse Hematopoietic Progenitors’ Formation Using Time-lapse Microscopy and Image Analysis. Bio-protocol 9(1): e3137. DOI: 10.21769/BioProtoc.3137.
- Bergiers, I., Andrews, T., Vargel Bolukbasi, O., Buness, A., Janosz, E., Lopez-Anguita, N., Ganter, K., Kosim, K., Celen, C., Itir Percin, G., Collier, P., Baying, B., Benes, V., Hemberg, M. and Lancrin, C. (2018). Single-cell transcriptomics reveals a new dynamical function of transcription factors during embryonic hematopoiesis. Elife 7: e29312.
Category
Developmental Biology > Cell growth and fate > Differentiation
Stem Cell > Embryonic stem cell > Cell-based analysis
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










