- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In planta Transcriptome Analysis of Pseudomonas syringae
Published: Vol 8, Iss 17, Sep 5, 2018 DOI: 10.21769/BioProtoc.2987 Views: 9341
Reviewed by: Zhibing LaiChao-Jan LiaoReza Sohrabi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Mungbean Yellow Mosaic India Virus (MYMIV)-infection, Small RNA Library Construction and Deep Sequencing for MicroRNA Identification in Vigna mungo
Anirban Kundu [...] Genotypic Technology
Oct 20, 2016 11115 Views

Isolation of Nuclei in Tagged Cell Types (INTACT), RNA Extraction and Ribosomal RNA Degradation to Prepare Material for RNA-Seq
Mauricio A. Reynoso [...] Kaisa Kajala
Apr 5, 2018 16204 Views
Abstract
Profiling bacterial transcriptome in planta is challenging due to the low abundance of bacterial RNA in infected plant tissues. Here, we describe a protocol to profile transcriptome of a foliar bacterial pathogen, Pseudomonas syringae pv. tomato DC3000, in the leaves of Arabidopsis thaliana at an early stage of infection using RNA sequencing (RNA-Seq). Bacterial cells are first physically isolated from infected leaves, followed by RNA extraction, plant rRNA depletion, cDNA library synthesis, and RNA-Seq. This protocol is likely applicable not only to the A. thaliana–P. syringae pathosystem but also to different plant-bacterial combinations.
Keywords: RNA-SeqBackground
Plants have evolved innate immune systems to fend off pathogen attack. Molecular mechanisms of pathogen recognition and immune signaling pathways have been intensively studied over the past decades. However, how plant immunity affects pathogen metabolism to inhibit pathogen growth is scarcely understood often because of the difficulty in profiling pathogen responses in planta. In case of bacterial pathogens, transcriptome profiling inside plant leaves is difficult to study because the amount of bacterial mRNA is much lower than that of plants; and it is particularly challenging at an early stage of infection due to the low population density of bacteria in plants. To overcome this limitation, we established a method for isolating bacteria from the infected plant leaves and profiling bacterial transcriptome with RNA-Seq. This method has been successfully used for profiling the transcriptome of the model bacterial pathogen Pseudomonas syringae pv. tomato DC3000 in the model plant Arabidopsis thaliana in various conditions (Nobori et al., 2018).
Materials and Reagents
- 50 ml Falcon tubes (Corning, Falcon®, catalog number: 352070 )
- 1.5 ml Eppendorf Safe-Lock tubes (Eppendorf, catalog number: 0030120086 )
- Pipette tips, 1,000 μl and 10 μl volumes (Corning, DeckWorksTM, catalog numbers: 4124 and 4120 )
- Sterile Serological Pipet, 25 ml (Corning, catalog number: 4251 )
- 1 ml needleless syringe (BD, Luer-LokTM, catalog number: 303172 )
- 4 mm stainless steel balls (Mühlmeier)
- 6 μm filter mesh (Bückmann, catalog number: 20000796 )
- 30-32 days old Arabidopsis thaliana plants
- Pseudomonas syringae pv. tomato DC3000 (Pto)
- 100% ethanol (VWR International, catalog number: 20821.321 )
- Chloroform (Carl Roth, catalog number: 4432 )
- peqGOLD TriFast (VWR, PeqLab, catalog number: 30-2010 )
- Phenol solution pH 8.0 (Sigma-Aldrich, catalog number: P4557 )
- Nuclease-Free Water (not DEPC-Treated) (Thermo Fisher Scientific, InvitrogenTM, Ambion®, catalog number: AM9932 )
- Tris(2-carboxyethyl)phosphine (TCEP) (Sigma-Aldrich, catalog number: C4706 )
- RNAqueous®-Micro Total RNA Isolation Kit (Thermo Fisher Scientific, InvitrogenTM, Ambion®, catalog number: AM1931 )
- TURBO DNA-free Kit (Thermo Fisher Scientific, InvitrogenTM, Ambion®, catalog number: AM1907 )
- Ribo-Zero rRNA Removal Kit (Plant) (Illumina, catalog number: MRZPL1224 )
- RNeasy Mini Kit (QIAGEN, catalog number: 74104 )
- Ovation® Complete Prokaryotic RNA-Seq DR Multiplex Systems (NuGEN Technologies, catalog number: 0326-32 )
- QIAGEN-tip 500 (QIAGEN, catalog number: 10063 )
- Liquid nitrogen
- NaOH (Carl Roth, catalog number: 6771 )
- Bacterial isolation buffer (see Recipes)
Equipment
- Eppendorf Reference Pipette (Eppendorf, 100-1,000 μl)
- Gilson PIPETMAN Classic pipette (Gilson, model: P20, catalog number: F123600 )
- Electric pipette (BrandTech Scientific, model: accu-jet® pro, catalog number: 26330 )
- Refrigerated centrifuge (Eppendorf, models: 5417 R and 5810 R )
- Roller mixer SRT1 (Stuart Scientific, model: SRT1 )
- Vortex mixer (Scientific Industries, model: Vortex-Genie 2 , catalog number: SI-0236)
- Shaker
- Thermal cycler (DNA Engine Tetrad 2 Peltier Thermal Cycler, Bio-Rad, model: Tetrad 2 )
- NanoDropTM One/OneC Microvolume UV-Vis Spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM OneC , catalog number: ND-ONE-W)
- -80 °C freezer
- Autoclave
- Fume hood
- Forceps (Fine Science Tools, model: Dumont #5, catalog number: 11251-10 )
- Plastic cutter
- Scissors
- Illumina HiSeq-3000 sequencer
Software
- BowTie2
- Python package HTSeq
Procedure
The overview of the procedure is shown in Figure 1.![]()
Figure 1. Workflow summary of in planta bacterial transcriptome analysis. The figure is adopted from Nobori et al. (2018), PNAS. Leaves infected with P. syringae are crushed and incubated in the bacterial isolation buffer, followed by filtering out large plant tissues. Then the flow through is centrifuged to separate bacterial cells from plant cells. Bacterial cells are harvested and subjected to RNA extraction, cDNA library preparation, and RNA-seq.
- Plant growth condition
Plants were grown in a chamber at 22 °C with a 10-h light period and 60% relative humidity for 24 d and then in another chamber at 22 °C with a 12-h light period and 60% relative humidity. Light intensity was around 120 μmol/m2/sec. Thirty to thirty-two days old A. thaliana plants old were used. - Bacterial isolation (Day 1)
Note: Use protective equipment while touching the frozen tubes.- Infiltrate bacterial suspension (OD600 = 0.5) to A. thaliana leaves (4 leaves/plant x 20 plants, 80 leaves in total, approximately 3 g) from the abaxial side with a 1 ml needle-less syringe. Make sure the entire leaf area infiltrated with the bacteria.
- Six hours after infiltration, collect the 80 leaves with forceps into a 50 ml Falcon tube containing 8-10 stainless steel balls (4 mm; sterilized by washing with ethanol and air dried). Flash-freeze with liquid nitrogen. Samples can be stored at -80 °C.
- Shake thoroughly by hand and crush the leaves into small pieces (Figure 2). Ensure that the samples do not thaw during crushing.

Figure 2. Crushed leaf sample before incubation in the bacterial isolation buffer - Add 30 ml ice-cold bacterial isolation buffer in a fume hood and vortex immediately for 10 sec followed by shaking vigorously by hand for 10 sec. Incubate the tubes for 20 h with shaking at 33 rpm using Roller mixer SRT1 at 4 °C.
- Bacterial isolation (Day 2)
Note: Steps C1-C7 and D1-D12 should be done in a fume hood.- Equip a 50 ml Falcon tube with a filter holder (handmade from a QIAGEN-tip 500 column; the upper half was cut by a plastic cutter; autoclaved before using) and a single layer of 6 µm filter mesh (cut into 12 cm x 15 cm size, and then autoclaved) (Figure 3). Trim away the excess filter mesh with scissors so that the cap can fit the tube.
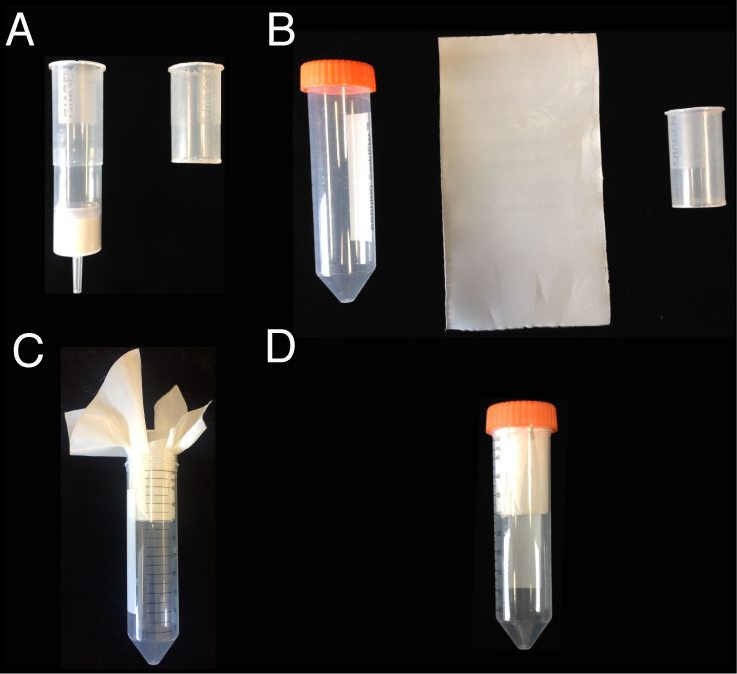
Figure 3. Filter and filter holder equipped to a 50 ml Falcon tube. A. A QIAGEN-tip 500 column is cut to make a filter holder. B. A 50 ml Falcon tube, a 6 µm filter mesh (12 cm x 15 cm), and the filter holder. C. Equip the 50 ml Falcon tube with the filter holder and the filter mesh. D. Trim away the excess filter mesh so that the cap can fit the tube. - Apply the sample (15 ml; without stainless steel balls) to the column and filter it by centrifuging at 1,300 x g for 10 sec (or until all the liquid goes through) at 4 °C. While keeping the flow-through in the tube, apply the rest of the sample (15 ml) to the column and repeat the filtering.
- Remove the filter and the filter holder. Centrifuge the flow-through for 20 min at 3,200 x g at 4 °C. Discard the supernatant with an electronic pipette equipped with a 25 ml serological pipette. You may leave some liquid (100 μl) to avoid the sample loss.
- Add 900 μl of ice-cold bacterial isolation buffer and resuspend the pellet by vortexing, then transfer it to a new 1.5 ml tube on ice by pipetting. Make sure that the pellet is completely resuspended.
- Centrifuge for 20 min at 2,300 x g at 4 °C. You will see two-layered pellet: white on the top and green at the bottom (Figure 4).
Note: Sometimes the upper part of the tube gets pale green. In this case, you may remove the green part by pipetting with a small amount of the liquid phase and discard it to avoid a potential plant contamination.
Figure 4. A two-layered pellet consists of plants (bottom, green) and bacteria (top, white)
- Equip 20 μl Gilson pipette with 10 μl filter tip (with 20 μl capacity) and set the volume to 18 μl. Carefully resuspend only the top white layer by pipetting up and down close to the surface of the pellet. When the buffer gets cloudy by the bacterial cells, transfer the buffer (approximately 1,000 μl) to a new 1.5 ml tube on ice using 1,000 μl pipette. Then, carefully add 1 ml of new ice-cold bacterial isolation buffer without disrupting the pellet by tiling the tube. Repeat resuspending and collecting bacterial cell suspensions in new 1.5 ml tubes (total three times or until the white layer is completely removed).
Notes:- You may use larger tips that fit the pipette, but smaller tips make it easier to resuspend the bacterial layer without breaking the plant layer.
- If the layer of plant tissues is collapsed and mixed with the layer of bacterial cells, you can recover the bacterial layer by completely resuspending the entire pellet by vortexing and then repeating the centrifugation step (Step C5).
- Centrifuge the bacterial suspensions for 2 min at 10,000 x g to harvest bacterial cells. Discard the supernatant by pipetting.
- Equip a 50 ml Falcon tube with a filter holder (handmade from a QIAGEN-tip 500 column; the upper half was cut by a plastic cutter; autoclaved before using) and a single layer of 6 µm filter mesh (cut into 12 cm x 15 cm size, and then autoclaved) (Figure 3). Trim away the excess filter mesh with scissors so that the cap can fit the tube.
- RNA extraction
- Add 1 ml of peqGOLD TriFast per sample to the pellets. For example, if you have 3 tubes after Step B6, add 333 μl of peqGOLD TriFast to each tube, vortex, and merge them in one tube. Samples can be stored at -80 °C.
- Add 200 μl of chloroform per 1 ml of peqGOLD TriFast, vortex, and leave the tube for 2 min at room temperature (RT).
- Centrifuge for 15 min at 12,000 x g at 4 °C. Take the aqueous part (typically 400-500 μl) into a new 1.5 ml Eppendorf tube by pipetting.
- Isolate RNA using RNAqueous®-Micro Total RNA Isolation Kit (Invitrogen, Ambion®) according to the manufacturer's protocol with some modifications (see below).
- Preheat Elution Solution to 75 °C.
- Add half-volume of 100% ethanol to the sample and vortex briefly.
- Load the sample (up to 150 μl) onto a Micro Filter Cartridge Assembly. Centrifuge for 10 sec at 12,000 x g at RT. Repeat this step until all of the samples are loaded. Discard flow-through.
- Add 180 μl of Wash Solution 1 to the Micro Filter Cartridge. Centrifuge for 10 sec at 12,000 x g at RT.
- Add 180 μl of Wash Solution 2/3 to the Micro Filter Cartridge. Centrifuge for 10 sec at 12,000 x g at RT.
- Repeat Step C9 one more time.
- Replace the Micro Filter Cartridge into a fresh Collection Tube. Centrifuge for 1.5 min at 12,000 x g at RT to remove residual fluid and dry the filter.
- Replace the Micro Filter Cartridge into a new 1.5 ml Eppendorf tube. Elute RNA with 15 μl of preheated Elution Solution. Repeat elution one more time into the same tube (30 μl in total).
- Perform DNase treatment with TURBO DNA-free kit (Ambion®) according to the manufacturer's protocol.
- Check the RNA concentration with NanoDrop. Typical RNA concentration is 100-200 ng/μl.
- Plant-derived rRNA depletion
Follow the protocol of Ribo-Zero rRNA Removal Kit (Plant). Use RNeasy Mini Kit (QIAGEN) for RNA purification as described in the manufacturer's protocol. Typical RNA yield is 100-400 ng. - Library preparation and sequencing
- cDNA library is synthesized using Ovation® Complete Prokaryotic RNA-Seq DR Multiplex Systems (NuGEN) according to the manufacturer's protocol with some modifications.
IMPORTANT: Input 10 ng of bacteria-enriched RNA, although the kit recommends 100-500 ng. This is because you cannot always obtain this much enriched RNA. Keep the RNA input amount consistent. - Sequence the library with an Illumina HiSeq sequencer.
- cDNA library is synthesized using Ovation® Complete Prokaryotic RNA-Seq DR Multiplex Systems (NuGEN) according to the manufacturer's protocol with some modifications.
Data analysis
Mapping and counting of Illumina reads for transcriptome:
- Sequence reads were mapped on Pto genome (available at http://pseudomonas.com/) using BowTie2 (Langmead and Salzberg, 2012). Sequence reads with the phred quality score greater than 33 were taken as an input (Command: --phred33). We use default settings for the other parameters.
- Mapped reads were counted with the Python package HTSeq (Anders et al., 2015) with default settings. The gene annotation file (GFF3 file) of Pto is available at http://pseudomonas.com/.
Notes
- Both phenol and chloroform are toxic. Handle them in a fume hood and wear proper protection.
- In Step D14, the quality of RNA tends to be variable and often low. However, this did not cause problems in data reproducibility and accuracy of RNA-Seq measurement in our previous experiments (Nobori et al., 2018).
Recipes
- Bacterial isolation buffer
Nuclease-Free Water
25 mM TCEP (tris(2-carboxyethyl)phosphine), pH 4.5 adjusted with 10 N NaOH
9.5% Ethanol
0.5% Phenol solution
Note: Prepare it fresh every time.
Acknowledgments
This protocol has been adapted from a previously published paper (Nobori et al., 2018). This work was supported by the Max Planck Society and Deutsche Forschungsgemeinschaft Grant SFB670 (to K.T.) and a predoctoral fellowship from the Nakajima Foundation (to T.N.).
Competing interests
The authors declare no conflicts of interest or competing interests.
References
- Nobori, T., Velasquez, A. C., Wu, J., Kvitko, B. H., Kremer, J. M., Wang, Y., He, S. Y. and Tsuda, K. (2018). Transcriptome landscape of a bacterial pathogen under plant immunity. Proc Natl Acad Sci U S A 115(13): E3055-E3064.
- Anders, S., Pyl, P. T. and Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2): 166-169.
- Langmead, B. and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4): 357-35
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Nobori, T. and Tsuda, K. (2018). In planta Transcriptome Analysis of Pseudomonas syringae. Bio-protocol 8(17): e2987. DOI: 10.21769/BioProtoc.2987.
Category
Plant Science > Plant molecular biology > RNA > RNA sequencing
Microbiology > Microbe-host interactions > Bacterium
Molecular Biology > RNA > RNA sequencing
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










