- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Autophagic Activity Using ATG8 Lipidation Assay in Arabidopsis thaliana
Published: Vol 8, Iss 12, Jun 20, 2018 DOI: 10.21769/BioProtoc.2880 Views: 8310
Reviewed by: Zhibing LaiYasin DagdasAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantitative Estimation of Auxin, Siderophore, and Hydrogen Cyanide Production in Halo and Drought-Tolerant Bacterial Isolates for Cucumber Growth
Zeinab Fotoohiyan and Ali Salehi Sardoei
Oct 5, 2025 1342 Views

Reproducible Emu-Based Workflow for High-Fidelity Soil and Plant Microbiome Profiling on HPC Clusters
Henrique M. Dias [...] Christopher Graham
Jan 20, 2026 403 Views
Abstract
As a fundamental metabolic pathway to degrade and recycle cellular cargos, autophagy is highly induced upon stress, starvation and senescence conditions in plants. A double-membrane structure named autophagosome will form during this process for cargo sequestration and delivery into the vacuole.
A number of regulators have been characterized in plants, including the autophagy-related (ATG) proteins and other plant-specific proteins. Among them, ATG8 will undergo a lipidation process to become a membrane-bound ATG8-phosphatidylethanolamine form and mark the growing autophagosomal membrane as well as the completed autophagosome. Therefore, ATG8 has been regarded as a marker for autophagosomes; and biochemical detection of the membrane-associated form of ATG8 is used as one of the principal methods for measurement of autophagic activity. Here, we describe an ATG8 lipidation assay for detection of the ATG8-PE form using Arabidopsis thaliana seedlings.
Background
Autophagy is an essential metabolic process which mediates the bulk degradation of the damaged organelles and unwanted cellular contents. During autophagy, a double-membrane structure called autophagosome will form and deliver the cargos into the vacuole for degradation. The autophagy-related (ATG) proteins are required to regulate the autophagic activity (Liu and Bassham, 2012). Among them, two conjugation systems, including ATG8 conjugate and ATG5-ATG12 conjugate, are involved for autophagosome formation. Upon autophagic induction, the ATG5-ATG12 conjugate forms and functions as an E3-like enzyme to promote ATG8 lipidation for binding to the phosphatidylethanolamine (PE) on the autophagosome membrane (Ohsumi, 2001). Although ATG8-PE on the outer membrane will be recycled before the autophagosome fusion with the vacuole, ATG8-PE on inner membrane will traffic together with the cargo into the vacuole for degradation. Thus, the amount of ATG8-PE usually correlates with the number of punctate ATG8-positive structures as well as autophagic activity (Mizushima et al., 2010). Particularly, due to the high hydrophobicity of ATG8-PE, ATG8-PE migrates faster than ATG8 in SDS-PAGE gel, though the actual molecular weight of ATG8-PE is larger than the unconjugated ATG8 (Mizushima and Yoshimori, 2007). Accordingly, the amount of ATG8-PE from cell membrane fraction (CM) can be detected by immunoblotting with ATG8 antibodies. For example, in Arabidopsis atg5 mutant, the level of ATG8-PE is severely impaired upon autophagic induction, whereas no autophagosome structures labeled by ATG8 are formed (Chung et al., 2010). Therefore, biochemical detection of the ATG8 lipidation can serve as a useful method to access the autophagic activity when combined with different treatments, which has been applied in our previous study as well as others related to plant autophagy (Chung et al., 2010; Suttangkakul et al., 2011; Li et al., 2014; Zhuang et al., 2017). Here, we describe the protocol for ATG8 lipidation detection by ultracentrifuge separation of the membrane and cytosol fractions using acibenzolar-S-methyl (BTH)-treated seedlings (Zhuang et al., 2017).
Materials and Reagents
- 10 µl pipette tips (Thermo Fisher Scientific, catalog number: 3510 )
- 200 µl pipette tips (Wolf Laboratories, catalog number: 2100.YN )
- 1,000 µl pipette tips (Thermo Fisher Scientific, catalog number: 3580 )
- 1.5 ml microcentrifuge tubes (Corning, Axygen®, catalog number: MCT-150-C )
- PVDF membrane
- X-ray film (Advansta, catalog number: L-07014-100 )
- 5-day-old seedlings
- MS salt (Caisson, catalog number: MSP01-50LT )
- UltraPureTM Sucrose (Thermo Fisher Scientific, InvitrogenTM, catalog number: 15503022 )
- BTH (Acibenzolar-S-methyl) (Sigma-Aldrich, catalog number: 32820 )
- Methanol (VWR, BDH, catalog number: 10158 )
- Liquid nitrogen
- PIC (Protease Inhibitor Mixture) (Roche Diagnostics, catalog number: 11873580001 )
- Tris Base (Caisson, catalog number: T041-1KG )
- NaCl (Alfa Aesar, USB, catalog number: J21618 )
- EDTA (Alfa Aesar, USB, catalog number: J15701 )
- SDS (Sodium dodecyl sulfate) (Alfa Aesar, USB, catalog number: J75819 )
- Triton X-100 (GE Healthcare, Amerrsham, catalog number: 17-1315-01 )
- Glycerol ultrapure (Alfa Aesar, USB, catalog number: J16374 )
- Bromophenol Blue (Sigma-Aldrich, catalog number: B5525 )
- β-mercaptoethanol (Sigma-Aldrich, catalog number: M3148 )
- Urea (Alfa Aesar, USB, catalog number: J75826 )
- 40% Acrylamide/Bis Solution (Bio-Rad Laboratories, catalog number: 161-0148 )
- TEMED (Bio-Rad Laboratories, catalog number: 161-0801 )
- APS (Ammonium persulfate) (USB, catalog number: US76322 )
- Non-fat milk powder
- ATG8 antibody (Agrisera, catalog number: AS14 2769 )
- cFBPase (Agrisera, catalog number: AS04 043 )
- Secondary antibody (Anti-rabbit IgG peroxidase conjugate, Sigma-Aldrich, catalog number: A6154 )
- Precision Plus ProteinTM Dual Color Standards (Bio-Rad Laboratories, catalog number: 161-0374 )
- Sodium hydrogen carbonate (NaHCO3) (VWR, catalog number: 144-55-8 )
- Sodium carbonate (Na2CO3) (USB, catalog number: 21602 )
- Sodium phosphate monobasic monohydrate (NaH2PO4·H2O) (USB, catalog number: 20233 )
- Sodium phosphate dibasic dihydrate (Na2HPO4·2H2O) (Sigma-Aldrich, catalog number: 04272 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: 31248 )
- Tween-20 (Sigma-Aldrich, catalog number: 63158 )
- MS liquid medium (see Recipes)
- 10 mM BTH stock (see Recipes)
- 25x PIC (see Recipes)
- 10% (v/v) Triton X-100 (see Recipes)
- 1 M Tris-HCl stock (pH 6.8 or pH 7.4) (see Recipes)
- 0.5 M EDTA (pH 8.0) (see Recipes)
- 5x extraction buffer (see Recipes)
- 1x extraction buffer containing 1x PIC and 1% (v/v) Triton X-100 (see Recipes)
- 5x sample loading dye (see Recipes)
- 30% APS (see Recipes)
- 3x separation buffer (see Recipes)
- 5x stacking buffer (see Recipes)
- 15% SDS-PAGE gel with 6 M urea (see Recipes)
- 15% Urea separating gel
- 5% Stacking gel
- 15% Urea separating gel
- Running buffer (see Recipes)
- Transfer buffer (see Recipes)
- PBS (see Recipes)
- PBS-T (see Recipes)
Equipment
- Eppendorf Research® plus Pipette 0.5-10 µl (Eppendorf, catalog number: 3120000020 )
- Eppendorf Research® plus Pipette 10-100 µl (Eppendorf, catalog number: 3120000046 )
- Eppendorf Research® plus Pipette 100-1,000 µl (Eppendorf, catalog number: 3120000062 )
- Mortar and pestle
- Centrifuge (Eppendorf, model: 5430 )
- X-ray film cassette (Amersham Biosciences HypercassetteTM)
- Ultracentrifuge tube (7 x 20 mm)
- Ultracentrifuge (Beckman Coulter, model: OptimaTM MAX-XP )
- Western Blotting apparatus (Bio-Rad)
- Developer machine (Fujifilm FPM100A)
Procedure
- Incubate 0.2 g 5-day Arabidopsis thaliana seedlings in MS liquid medium with and without 100 µM BTH (10 mM stock in methanol) for 8 h (see Notes 1 and 2).
- Freeze the seedlings in the mortar using liquid nitrogen and grind the plants thoroughly.
- Add 2x extraction buffer containing PIC without detergent into the mortar on ice.
- After defrost slowly on ice, transfer the liquid to a 1.5 ml tube and centrifuge at 1,000 x g for 5 min, 4 °C.
- Transfer the supernatant to an ultracentrifuge tube (7 x 20 mm) and centrifuge at 100,000 x g for 45 min, 4 °C.
- Transfer the supernatant (Cell Soluble fraction, CS) into a labeled new Eppendorf tube and keep on ice.
- Wash the pallet (Cell Membrane fraction, CM) slightly with the 1x extraction buffer containing 1x PIC for 2-3 times.
Note: In this step, add the 1x extraction buffer slightly without touching the surface of the pallet, and the centrifuge and re-suspend are not required. The main purpose of this step is washing out the remaining CS liquid on the CM surface. - Resuspend the pallet using 1x extraction buffer containing 1x PIC and 1% (v/v) Triton X-100 to solubilize the membranes.
- Add the 5x sample loading dye to both of the CS and CM samples.
- Boil the samples at 100 °C for 10 min.
- Prepare the 15% SDS-PAGE gel with 6 M urea.
Note: Only add the urea in the separating gel. - Load the CM samples and run at 90 V for 3 h with 1x running buffer. Run for additional 30 min at 90 V after the blue band reaches the bottom of the gel (see Notes 3 and 4).
- Transfer the proteins from the gel to PVDF membrane at 55 V for 2.5 h with 1x transfer buffer.
- After blotting, soak the blotted membrane in 1x PBS with 5% milk powder for 1 h. Wash with PBS-T for 3 times.
- Incubate with ATG8 specific primary antibody (4 μg/μl in PBS-T) for 1 h. Wash with PBST for 3 times.
- Incubate with the secondary antibody (1:5,000 in PBS-T) for 1 h. Wash with PBS-T for 3 times.
- Add Western blotting detecting reagents onto the membrane, and expose it to the X-ray film or use an imaging machine. (Figure1)
Data analysis
As shown in Figure 1, in the wild-type (WT) background, the ATG8-PE from the CM sample increases after BTH induction, with a size about 12-15 kDa, indicating that autophagy is induced with the formation of autophagosomes. However, ATG8-PE is not detected in the ATG5 deficient mutant after autophagic induction, implying that autophagosome formation is inhibited. Differently, a higher level of ATG8-PE was detected in atg9 mutant, suggesting that autophagosome formation might be interrupted at a certain stage. There might be non-specific bands in the ATG8 antibody detection. Also, it should be pointed out that there are multiple isoforms of ATG8 in the Arabidopsis genome with different SDS-PAGE mobilities, resulting the detection of cross-reacting species with similar size to the ATG8-PE adducts and making the results contradictory (Chung et al., 2010). Therefore, it is critical to include both the WT and atg5 samples as the positive and negative controls respectively to identify the correct size of ATG8-PE, as atg5 mutant lacks ATG8-PE but accumulates a large amount of non-lipidated ATG8. Our further examinations under confocal and electron microscopy identified that abnormal tubules labeled by ATG8 accumulated in the atg9 mutant, thus demonstrating that ATG9 is required for autophagosome progression (Zhuang et al., 2017). Therefore, the ATG8 lipidation assay should combine with other approaches such as microscopy analysis to further assess autophagic activity.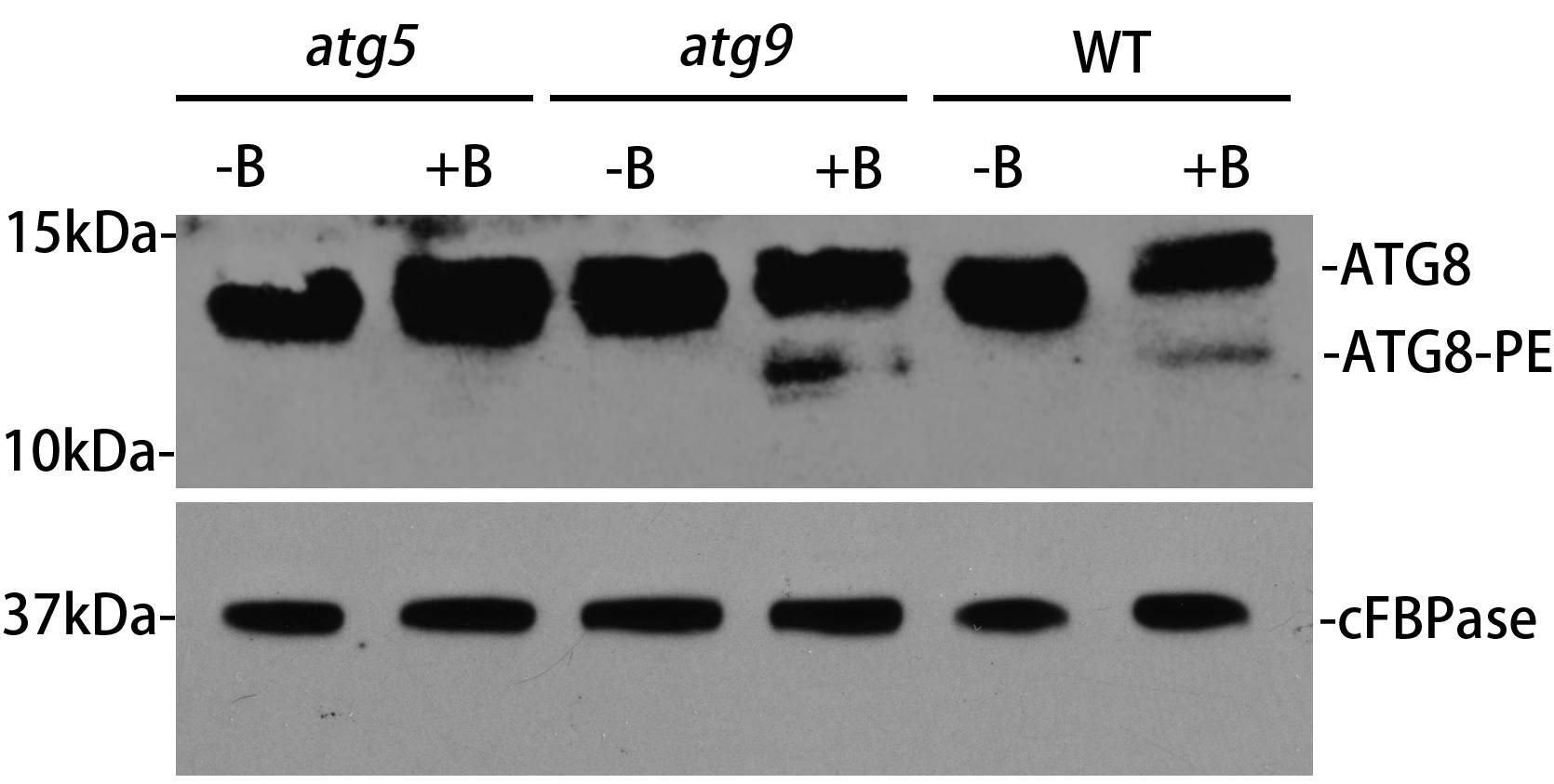
Figure 1. ATG8 lipidation detection in Arabidopsis thaliana wild-type and atg mutants. The 5-day-old wild-type, atg5 and atg9 seedlings were incubated in the MS liquid medium with or without BTH treatment (+B and -B) for 8 hours respectively. Crude extracts were subjected to ultracentrifuge to collect the cell membrane fraction (CM), followed by immunoblotting with plant ATG8 antibody. cFBPase represents the loading control with immunoblotting cFBPase antibodies.
Notes
- It is critical to make sure that the seedlings are in good condition without any stress before the autophagic induction, otherwise stressed seedlings will have induced autophagic activity, which may obscure the results.
- It is recommended to include known atg mutants defective in ATG8 lipidation (e.g., atg5 or atg7) as negative controls in the lipidation assay.
- When running the SDS-PAGE, the urea gel will generate a lot of heat and it should keep at a low voltage, so that the two forms of ATG8 can be well separated.
- ATG8-PE only occurs on the membrane fraction, therefore the CM samples are used for detecting the ATG8-PE adducts. The CS fraction with non-lipidated ATG8 could be included in the western blot as a control to distinguish the non-lipidated ATG8 and lipidated ATG8.
Recipes
- MS liquid medium (350 ml)
Adjust the pH to 5.7 (with KOH)Components Volume/Quantity MS salt 1.515 g Sucrose 3.5 g ddH2O up to 350 ml - 10 mM BTH stock (10 ml)
Components Volume/Quantity BTH 0.021 g Methanol 10 ml - 25x PIC (2 ml)
Components Volume/Quantity PIC one tablet ddH2O 2 ml - 10% (v/v) Triton X-100 (100ml)
Components Volume/Quantity Triton X-100 10 ml ddH2O 90 ml - 1 M Tris-HCl stock (pH 6.8 or pH 7.4) (100 ml)
Adjust pH to 6.8 or 7.4 with conc. HClComponents Volume/Quantity Tris Base 12.114 g ddH2O to 100 ml - 0.5 M EDTA (pH 8.0) (100 ml)
Adjust the pH to 8.0 with NaOH (~10 g of NaOH pellets) and add to 100 ml with ddH2OComponents Volume/Quantity EDTA 16.81 g ddH2O 80 ml - 5x extraction buffer (10ml)
Components Volume/Quantity 250 mM Tris-HCl (pH 7.4) 2.5 ml of 1 M Tris-HCl (pH 7.4) 750 mM NaCl 0.438 g 5 mM EDTA 0.1 ml of 0.5 M stock ddH2O to 10 ml - 1x extraction buffer containing 1x PIC and 1% (v/v) Triton X-100 (10 ml)
Components Volume/Quantity 1x extraction buffer 2.5 ml of 5x extraction buffer 1x PIC 0.40 ml of 25x PIC 1% (v/v) Triton X-100 1 ml of 10% (v/v) Triton X-100 ddH2O to 10 ml - Sample loading dye (5x, 50 ml)
Components Volume/Quantity 1 M Tris-HCl (pH 6.8) 12.5 ml SDS 5 g Glycerol 25 ml Bromophenol Blue 0.25 g β-mercaptoethanol 6.25 ml ddH2O to 50 ml - 30% APS (10 ml)
Components Volume/Quantity APS 3 g ddH2O 10 ml - 3x separation buffer (1 L)
Adjust the pH to 8.8 with conc. HCl (~10 ml)Components Volume/Quantity Tris base 136.2 g SDS 3 g ddH2O up to 1 L - 5x stacking buffer (500 ml)
Adjust the pH to 6.8 with conc. HCl (~20 ml)Components Volume/Quantity Tris base 37.85 g SDS 2.5 g ddH2O up to 500 ml - 15% SDS-PAGE gel with 6 M urea
- 15% Urea separating gel (10 ml)
Components Volume/Quantity Urea 3.6036 g Acry-bis (40%) 3.75 ml 3x separation buffer 3.33 mlAPS (30%) 20 μl TEMED 5 μl ddH2O up to 10 ml - 5% Stacking gel (5 ml)
Components Volume/Quantity Acry-bis (40%) 0.625 ml5x stacking buffer 1 ml APS (30%) 20 μl TEMED 5 μl ddH2O up to 5 ml
- 15% Urea separating gel (10 ml)
- Running buffer (10x, 1 L)
Components Volume/Quantity Tris Base 30.3 g Glycine 144 g SDS 10 g ddH2O up to 1 L - Transfer buffer (10x, 1 L)
Components Volume/Quantity NaHCO3 8.4 g Na2CO3 3.2 g ddH2O up to 1 L - PBS (10x, 2 L)
Adjust the pH to 7.4 with 10 N NaOHComponents Volume/Quantity NaCl 80 g NaH2PO4·H2O 2.3 g Na2HPO4·2H2O 13.9 g KCl 2 g ddH2O up to 2 L - PBS-T (1 L)
Components Volume/Quantity 1x PBS 1 L Tween-20 0.5 ml
Acknowledgments
This protocol was adapted from Zhuang et al., 2017. Fundings from the Research Grants Council of Hong Kong (G-CUHK402/15, G-CUHK403/17, CUHK14130716, CUHK14102417, C4011-14R, C4012-16E, C4002-17G and AoE/M-05/12) and the National Natural Science Foundation of China (31470294 and 31670179) support this work.
Competing interests
The authors declare no conflicts of interests.
References
- Chung, T., Phillips, A. R. and Vierstra, R. D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J 62(3): 483-493.
- Li, F., Chung, T. and Vierstra, R. D. (2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 26(2): 788-807.
- Liu, Y. and Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215-237.
- Mizushima, N., Yoshimori, T. and Levine, B. (2010). Methods in mammalian autophagy research. Cell 140(3): 313-326.
- Mizushima, N. and Yoshimori, T. (2007). How to interpret LC3 immunoblotting. Autophagy 3(6): 542-545.
- Ohsumi, Y. (2001). Ubiquitin and proteasomes: Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2(3): 211-216.
- Suttangkakul, A., Li, F., Chung, T. and Vierstra, R. D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23(10): 3761-3779.
- Zhuang, X., Chung, K. P., Cui, Y., Lin, W., Gao, C., Kang, B. H. and Jiang, L. (2017). ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci U S A 114(3): E426-E435.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Luo, M. and Zhuang, X. (2018). Analysis of Autophagic Activity Using ATG8 Lipidation Assay in Arabidopsis thaliana. Bio-protocol 8(12): e2880. DOI: 10.21769/BioProtoc.2880.
Category
Plant Science > Plant immunity > Host-microbe interactions
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










