- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Metal-tagging Transmission Electron Microscopy for Localisation of Tombusvirus Replication Compartments in Yeast
Published: Vol 8, Iss 8, Apr 20, 2018 DOI: 10.21769/BioProtoc.2822 Views: 7417
Reviewed by: David PaulMirko CorteseAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

General Maintenance and Reactivation of iSLK Cell Lines
Ariana C. Calderón-Zavala [...] Ekaterina E. Heldwein
Jun 5, 2025 1875 Views

Inducible HIV-1 Reservoir Reduction Assay (HIVRRA), a Fast and Sensitive Assay to Test Cytotoxicity and Potency of Cure Strategies to Reduce the Replication-Competent HIV-1 Reservoir in Ex Vivo PBMCs
Jade Jansen [...] Neeltje A. Kootstra
Jul 20, 2025 2457 Views

Assembly and Mutagenesis of Human Coronavirus OC43 Genomes in Yeast via Transformation-Associated Recombination
Brett A. Duguay and Craig McCormick
Aug 20, 2025 3033 Views
Abstract
Positive-stranded (+) RNA viruses are intracellular pathogens in humans, animals and plants. To build viral replicase complexes (VRCs) viruses manipulate lipid flows and reorganize subcellular membranes. Redesigned membranes concentrate viral and host factors and create an environment that facilitates the formation of VRCs within replication organelles. Therefore, efficient virus replication depends on the assembly of specialized membranes where viral macromolecular complexes are turned on and hold a variety of functions. Detailed characterization of viral replication platforms in cells requires sophisticated imaging approaches. Here we present a protocol to visualize the three-dimensional organization of the tombusvirus replicase complex in yeast with MEtal-Tagging Transmission Electron Microscopy (METTEM). This protocol allowed us to image the intracellular distribution of the viral replicase molecules in three-dimensions with METTEM and electron tomography. Our study showed how viral replicase molecules build replication complexes within specialized cell membranes.
Keywords: Metal-tagging transmission electron microscopyBackground
Replication of positive-stranded RNA viruses depends on the remodeling of cellular membranes. Intracellular membranes serve as a structural scaffold for VRC assembly, provide essential lipids and co-factors that modulate the activity of the viral replicase and protect the viral RNA from the antiviral defenses of the host (Miller and Krijnse-Locker, 2008; den Boon et al., 2010; Nagy and Pogany, 2011; Nagy, 2016). The architecture of replication organelles with active VRCs has been observed by electron microscopy. VRCs assemble in single membrane vesicles or ‘spherules’, tubulovesicular cubic membranes, double membrane vesicles (DMV) or planar oligomeric arrays (de Castro et al., 2013). Spherules are often observed in cells infected by RNA viruses. They form by invagination in a variety of organelles and have a narrow opening to the cytosol (den Boon et al., 2010).
Tomato bushy stunt virus (TBSV) is a small (+) RNA virus that belongs to the Tombusviridae, a family of viruses that infect plants. TBSV has recently emerged as a model virus to study viral replication and virus-host interactions using the yeast Saccharomyces cerevisiae as a model host (Nagy and Pogany, 2011). Studies of S. cerevisiae infected with plant viruses have facilitated the identification of numerous factors needed for viral replication (Nagy, 2008). Tombusviruses encode five proteins including two replication proteins, p92pol and p33. p92pol is the RNA-dependent RNA polymerase. The auxiliary protein p33 is an RNA chaperone that facilitates the recruitment of the viral RNA to the site of replication, in the cytosolic face of peroxisome membranes (McCartney et al., 2005; Jonczyk et al., 2007).
Understanding the biogenesis and functional architecture of VRCs in cell membranes is challenging and it requires sophisticated imaging techniques. Transmission electron microscopy (TEM) has contributed to our understanding of the architecture and organization of macromolecular assemblies in cells. However, methods to unambiguously identify proteins within the environment of the cell are lagging behind. In our lab, we have developed a new labeling method named METTEM from MEtal-Tagging Transmission Electron Microscopy. This method uses the metal-binding protein metallothionein (MT) as a genetically clonable tag for electron microscopy (Diestra et al., 2009; Risco et al., 2012). Mouse MT 1 is a small, 61-amino acid protein with 20 cysteine residues that bind gold atoms very efficiently. MT fused to a protein of interest and treated with gold salts, builds an electron-dense gold nanocluster of around 1 nm diameter, easily visualized by electron microscopy (Mercogliano and DeRosier, 2006 and 2007) (Figure 1). METTEM allows identification and localization of intracellular proteins with high specificity and exceptional sensitivity at molecular-scale resolution ( Diestra et al., 2009; Delebecque et al., 2011; Bouchet-Marquis et al., 2012; Risco et al., 2012; Barajas et al., 2014a; de Castro Martin et al., 2017; Fernandez de Castro et al., 2017).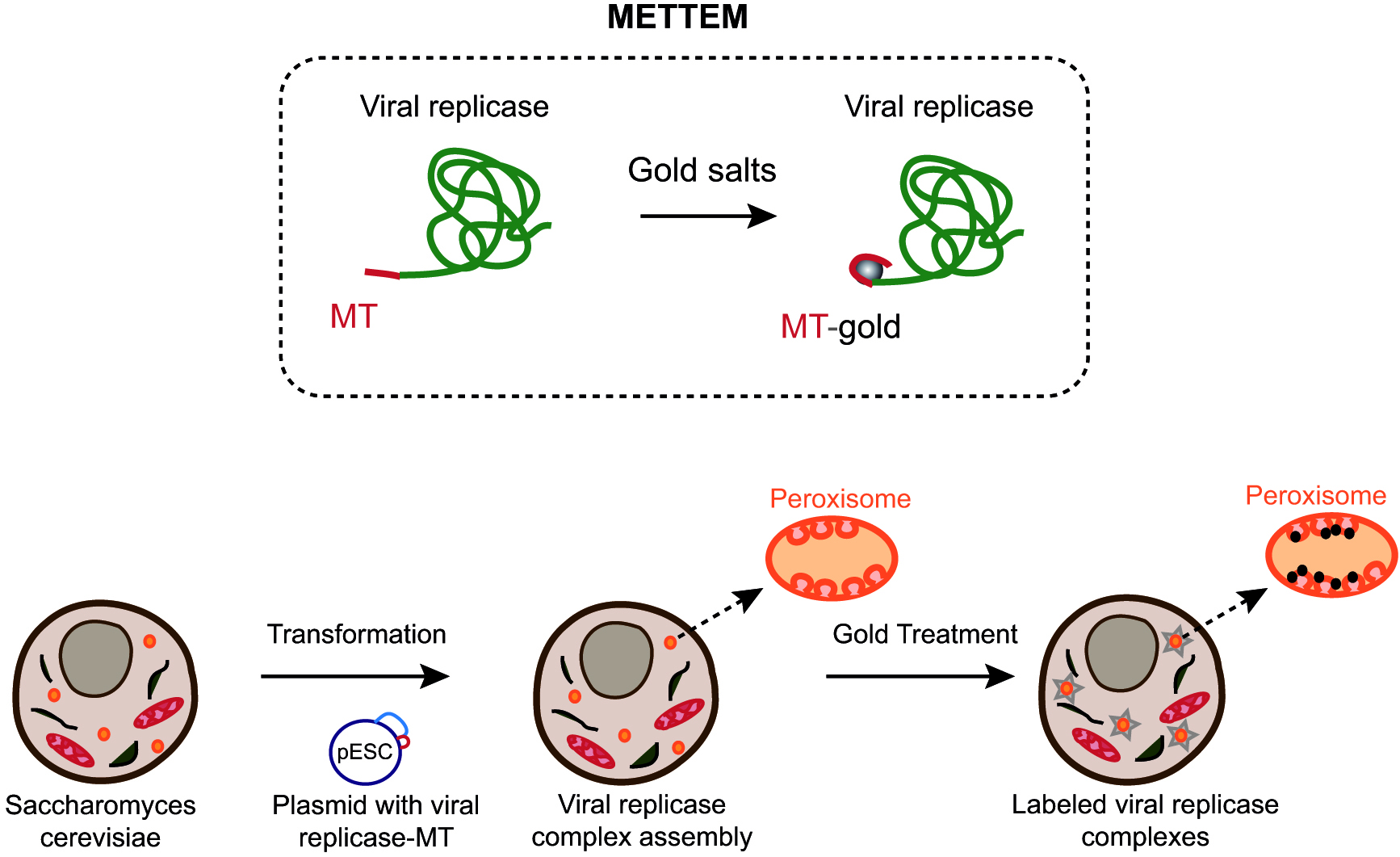
Figure 1. Imaging viral replicase complexes with METTEM. Viral replicase protein is fused with metallothionein (MT) and expressed in yeast cells (Fernandez de Castro et al., 2017). MT-tagged viral replicase molecules assemble VRCs in peroxisome membranes. Cells are incubated with gold salts in vivo and MT-gold-replicase molecules are visualized by electron microscopy.
Here we described a protocol to visualize TBSV replicase molecules in VRCs using METTEM (Figure 2). The combination of this technology with electron tomography allowed us to study the distribution of replicase molecules in the viral replication compartment in three-dimensions (3D). Due to the high sensitivity of the method we could distinguish different states of aggregation of the viral replicase molecules in situ. This methodology can be used to detect any protein of interest in different subcellular locations of bacteria, yeast and mammalian cells. Furthermore, one advantage of this electron microscopy approach is that it can be used to study many different viruses in a variety of cell types by visualizing the MT tag incorporated in either complete viral particles or their proteins. This method has revealed virus-induced structures not seen before, as reported for Rubella virus, Tombusvirus and influenza virus (Risco et al., 2012; de Castro Martin et al., 2017; Fernandez de Castro et al., 2014 and 2017). 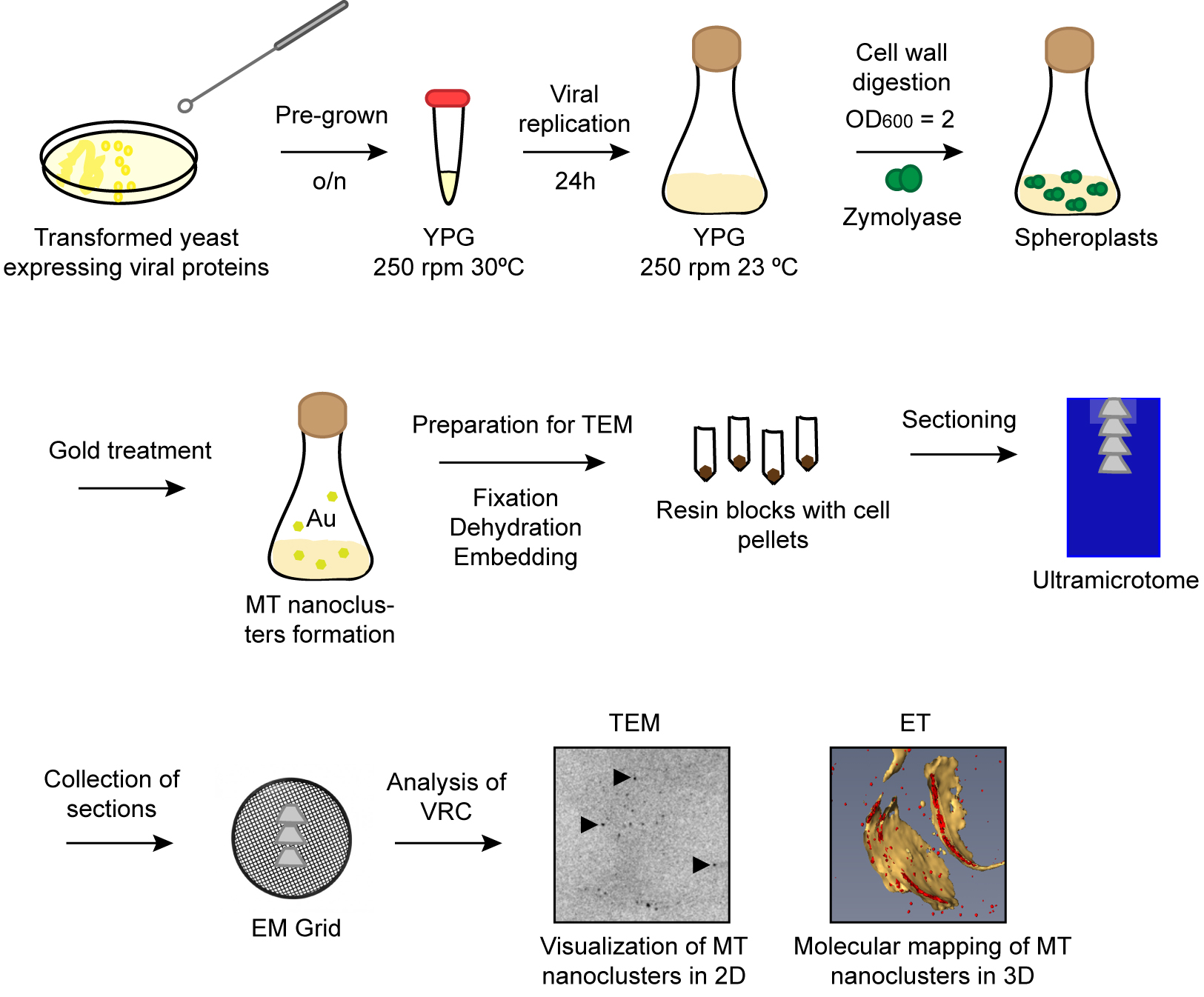
Figure 2. Schematic workflow of the protocol. Pre-grown transformed yeast cells are incubated overnight in YPG. Next day viral replication is induced during 24 h at 23 °C. Cells are treated with zymolyase to obtain spheroplasts. Spheroplasts are incubated with gold salts to build nanoclusters in MT tags. Cell pellets are dehydrated and embedded in resin. Serial sections are transferred to EM grids and imaged by TEM.
Materials and Reagents
- Pipette tips
- Perfect loop (Electron Microscopy Sciences, catalog number: 70945 )
- 50 ml disposable centrifuge tubes
- Eppendorf tubes
- Sterile transfer pipettes
- Serum Acrodisc® 37 mm syringe filter (Pall, catalog number: 4525 )
- Gelatin capsule size 1 6.5 mm diameter-0.50 ml (TAAB, catalog number: C089/1 )
- GEM® Single Edge Blades 3-Facet 0.009"/0.23 mm (AccuTec Blades, catalog number: 62-0179-0000 )
- Saccharomyces cerevisiae yeast strains RS453 (MATa ade2-1 his3, 15 leu2-3, 112 trp1-1 ura3 52) and pah1Δnem1Δ (SwissProt ID for Nem1 is P38757) (pah1Δ::TRP1nem1Δ::HIS3 derivative of RS453) (Choi et al., 2011; Barajas et al., 2014b)
- Zymolyase® 20T (Arthrobacter luteus) (Amsbio, catalog number: 120491-1 )
- Gold(III) chloride ≥ 99.99% (Sigma-Aldrich, catalog number: 379948 )
- Glutaraldehyde 50% (TAAB, catalog number: G015 )
- Ethanol Dry (Merck, catalog number: 1009901001 )
- LR-White Resin Medium Grade Acrylic resin (TAAB, catalog number: L012 )
- BactoTM yeast extract (BD, BactoTM, catalog number: 212750 )
- BactoTM peptone (BD, BactoTM, catalog number: 211677 )
- D-(+)-Glucose (Sigma-Aldrich, catalog number: G8270 )
- Agar (Sigma-Aldrich, catalog number: 05038 )
- D-(+)-Galactose (Sigma-Aldrich, catalog number: G0750 )
- Lithium acetate 99.95% (Sigma-Aldrich, catalog number: 517992 )
- ssDNA (Deoxyribonucleic acid sodium salt from salmon testes) (Sigma-Aldrich, catalog number: D1626 )
- PEG MW3350 (Polyethylene glycol) (Sigma-Aldrich, catalog number: P4338 )
- Trizma® hemisulfate (Sigma-Aldrich, catalog number: T8379 )
- 1,4-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: DTT-RO)
Manufacturer: Roche Diagnostics, catalog number: 10197777001 . - Yeast nitrogen base without amino acids (Sigma-Aldrich, catalog number: Y0626 )
- Yeast Synthetic Drop-out Medium Supplements without tryptophan (Sigma-Aldrich, catalog number: Y1876 )
- D-Sorbitol (Sigma-Aldrich, catalog number: S1876 )
- Tris (Base) (Norgen Biotek, catalog number: 28029 )
- Paraformaldehyde EM (TAAB, catalog number: P026 )
- NaOH
- 10x PBS
- 1,4-Piperazinediethanesulfonic acid, Piperazine-1,4-bis(2-ethanesulfonic acid), Piperazine-N,N’-bis(2-ethanesulfonic acid) PIPES (Sigma-Aldrich, catalog number: P6757 )
- 4-(2-Hydroxyethyl) piperazine-1-ethanesulfonic acid, N-(2-Hydroxyethyl) piperazine-N’-(2-ethanesulfonic acid) (HEPES) (Sigma-Aldrich, catalog number: H3375 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889 )
- YPD (see Recipes)
- YPG (see Recipes)
- Transformation mix (see Recipes)
- TSD reduction buffer (see Recipes)
- Spheroplast medium A (see Recipes)
- 4% paraformaldehyde (PFA) solution (see Recipes)
- PHEM solution, pH 6.9 (see Recipes)
Equipment
- 500 ml flask
- Pipettes
- Diamond Knife 45° DIATOME (Fedelco)
- QUANTIFOIL® R 3.5/1 100 Holey Carbon Films Grids Au 300 mesh (Quantifoil)
- Oven Memmert UN 55 (Memmert, model: UN55 ) (Genesys) equipped with a shaker
- Spectrophotometer 722N (Terra Universal, Laboratory Equipment, model: 722N )
- Centrifuge 5810 R (Eppendorf, model: 5810 R )
- Centrifuge miniSpin plus (Eppendorf, model: MiniSpin® plus )
- Ultrasonic Cleaner 1510 (Branson, model: 1510 )
- pH meter Basic 20 (HACH LANGE SPAIN, Crison, model: Basic 20 )
- Fume Hood (Flow-Tronic)
- Ultramicrotome (Leica Microsystems, model: Leica EM UC6 )
- Jeol JEM 1011 electron microscope operating at 100 kV (JEOL, model: JEM-1011 )
- FEI Tecnai G2 F20 (200 kV) electron microscope (FEI)
- Tecnai Spirit Twin (120 kV) electron microscope (FEI)
Software
- IMOD software
- Amira software
Procedure
- Transformation of Saccharomyces cerevisiae yeast strains
- Streak the yeast strain from a glycerol stock on a yeast extract peptone dextrose (YPD) agar plate. Grow cells at 30 °C for 1-2 days. Inoculate 10 ml YPD solution with a single colony with a loop. Grow culture overnight at 30 °C at 250 rpm (OD around 2).
- Dilute overnight culture to OD 0.2-0.3 in 50 ml YPD medium.
Note: Use a 500 ml flask. - Grow cells for 3-4 h at 30 °C at 250 rpm.
- Centrifuge cells at 3,590 x g for 5 min at room temperature. Use 50 ml disposable centrifuge tubes.
- Discard supernatant and add 1 ml of sterile dH2O, vortex gently, then fill up tube with sterile dH2O.
- Centrifuge cells at 3,590 x g for 5 min. Use 50 ml disposable centrifuge tubes.
- Discard supernatant and resuspend pellet in 1 ml of 100 mM lithium acetate.
- Transfer cells into an Eppendorf tube and centrifuge for 2 min at 3,590 x g. Discard supernatant and dissolve the cell pellet in 0.4 ml of 100 mM lithium acetate. Incubate at room temperature for 10-15 min.
Note: Use a sterile transfer pipette. - Add 1-5 μl plasmid DNA (5-10 μg) to each fresh Eppendorf tube.
- Make up the master mix for transformation (see Recipes).
- Add 0.4 ml transformation mix to each Eppendorf tube containing the plasmids. Vortex gently for 2 sec to mix components.
Note: Make sure that yeast cells are well resuspended. - Incubate cells at 30 °C (no shaking) for 30 min.
- Incubate cells at 42 °C (no shaking) for 40 min.
- Centrifuge cells at 570 x g for 2 min. Discard supernatant and add 100 μl sterile dH2O. Disperse pellet by gently pipetting up and down, and dispense cells on agar plates with the appropriate selection medium. Incubate plates at 30 °C for 2-3 days.
Note: Try to spread cells with few strokes.
- Streak the yeast strain from a glycerol stock on a yeast extract peptone dextrose (YPD) agar plate. Grow cells at 30 °C for 1-2 days. Inoculate 10 ml YPD solution with a single colony with a loop. Grow culture overnight at 30 °C at 250 rpm (OD around 2).
- Cell culture
- Pre-grow yeast in 2 ml of yeast extract peptone galactose (YPG) and incubate overnight at 30 °C and 250 rpm.
- Culture 50 ml of yeast cells in an Erlenmeyer for 24 h in YPG at 23 °C and 250 rpm.
Note: These conditions are necessary to induce and maintain viral replication. - Measure and control optical density at 600 nm (OD600) and when OD600 is around 2 harvest cells by centrifuging for 5 min, 4,000 x g, room temperature, and carefully remove supernatant.
- Resuspend cells at 5 to 10 OD600 units/ml in Tris-sulfate with dithiothreitol (DTT) (TSD) reduction buffer and incubate for 10 min at room temperature.
Notes:- DTT facilitates cell wall digestion by breaking disulfide bonds and making β-glucan linkages more accessible to the β-glucanase activity present in the zymolyase.
- The results of the experiment will change if cells are processed at different OD.
- DTT facilitates cell wall digestion by breaking disulfide bonds and making β-glucan linkages more accessible to the β-glucanase activity present in the zymolyase.
- Incubate yeast cells with 0.1 mg/ml zymolyase 20T for 10 min at 30 °C to obtain spheroplasts
Notes:- The optimal zymolyase concentration and digestion time depend on yeast strains and growth conditions. Setting zymolyase treatment is critical for an adequate preservation of cell ultrastructure.
- Before zymolyase treatment, it is important to sonicate cells for 5 sec in an Ultrasonic cleaner to avoid yeast aggregation.
- The optimal zymolyase concentration and digestion time depend on yeast strains and growth conditions. Setting zymolyase treatment is critical for an adequate preservation of cell ultrastructure.
- Centrifuge spheroplasts at 280 x g for 5 min and remove supernatant.
- Wash 3 times with ice-cold spheroplast medium A.
- Pre-grow yeast in 2 ml of yeast extract peptone galactose (YPG) and incubate overnight at 30 °C and 250 rpm.
- Gold treatment and fixation
- Incubate live spheroplasts with 2 mM HAuCl4 in spheroplast medium A for 75 min at room temperature in the dark.
Notes:- To prepare the HAuCl4 solution, dissolve in distilled and sterile water protected from light.
- MT binds gold atoms and builds a nanocluster visible by TEM.
- It is important to adjust the gold treatment conditions due to the potential toxicity of gold salts in eukaryotic cells.
- To prepare the HAuCl4 solution, dissolve in distilled and sterile water protected from light.
- Wash cells with spheroplast medium A.
- Fix cells with 4% paraformaldehyde and 0.2% glutaraldehyde in PHEM solution for 1 h at room temperature.
- After fixation wash spheroplasts 3 times with PHEM.
- Incubate live spheroplasts with 2 mM HAuCl4 in spheroplast medium A for 75 min at room temperature in the dark.
- Embedding
- Dehydrate cells in 10 min steps, with increasing concentrations of 1 ml ethanol (30, 50, 70, 90% and twice in 100%) at 4 °C. Centrifugation between steps is not necessary.
- Incubate cells in mixtures of ethanol–LR-White resin (2:1, 1:1, 1:2) on a rocking shaker for 1 h each at room temperature and protected from light.
Note: Do not expose LR-White resin to light and manipulate inside a well ventilated hood. - Remove cells from the ethanol-resin mix and place them in 100% resin for 24 h.
- Polymerize samples in gelatin capsules for 48 h at 60 °C.
- Dehydrate cells in 10 min steps, with increasing concentrations of 1 ml ethanol (30, 50, 70, 90% and twice in 100%) at 4 °C. Centrifugation between steps is not necessary.
- Ultramicrotomy
- Remove gelatin capsules and trim samples with a razor blade to make a trapeze.
- Collect 50-60 nm ultrathin sections on 300-mesh Quantifoil holey carbon grids.
- Observe sections under a transmission electron microscope at 30,000-50,000x nominal magnifications.
Note: Sections are observed by TEM without staining to avoid masking the small MT-gold nanoclusters.
- Remove gelatin capsules and trim samples with a razor blade to make a trapeze.
- Electron tomography and image processing
- Obtain semi-thick sections (~300 nm) and collect them on Quantifoil grids.
- Acquire tilt series automatically at 1.5° increments over an angular range of -60° to +60° on FEI Tecnai G2 F20 and Tecnai Spirit Twin microscopes with an accelerating voltage of 200 and 120 kV, respectively.
- Use IMOD software to align tilt series and for tomographic reconstruction.
- For tomogram segmentation and 3D reconstruction, Amira software is used.
Note: Noise reduction and automated segmentation software helps to highlight membrane visualization.
- Obtain semi-thick sections (~300 nm) and collect them on Quantifoil grids.
Data analysis
We have used the method METTEM to visualize the Tombusvirus replicase p33 in yeast cells. Electron microscopy imaging in Figure 3 showed the precise location of p33–MT-gold molecules in a membranous compartment compatible with peroxisome-derived multivesicular body (MVB), which is the Tombusvirus replication organelle in plant and yeast cells (Barajas et al., 2014a). With this approach, we could detect p33-metallothionein-gold nanoparticles in ER membranes inside the replication platform (Figures 3A and 3B).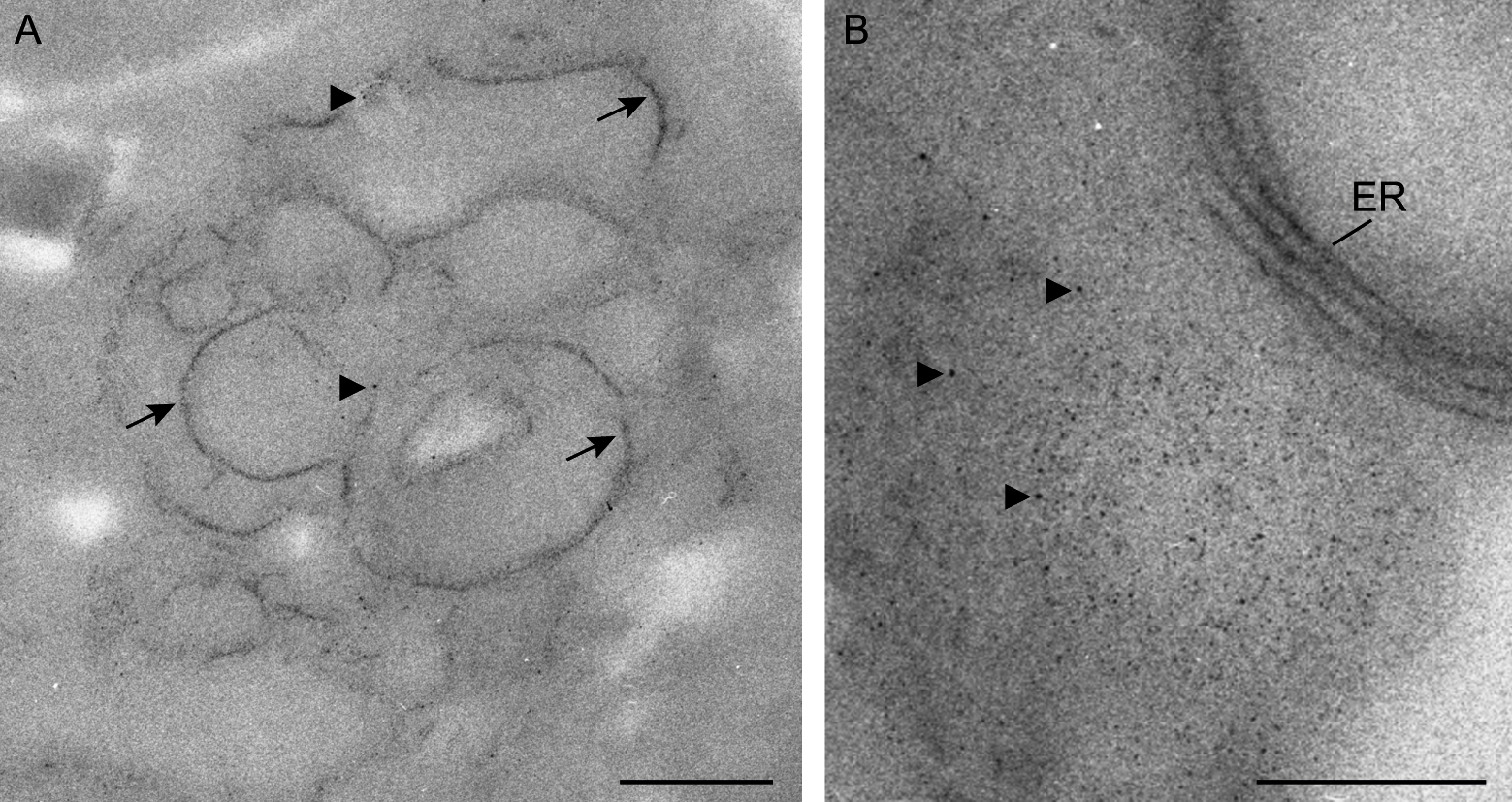
Figure 3. Visualization of p33–MT-gold molecules by METTEM. Sections were not stained in order to avoid masking the MT-gold nanoclusters. A. Inside the tombusvirus replication compartment p33–MT-gold nanoparticles (~1 nm) delineate membranes (arrows). Single MT clusters are marked with arrowheads. B. Tombusvirus replication platform with p33–MT-gold nanoclusters inside (arrowheads) and in the surrounding endoplasmic reticulum (ER) membranes. Scale bars = 200 nm.
For more precise details, we studied the viral replication organelles in three dimensions with electron tomography (Figure 4). Tomograms showed the internal organization of p33–MT-gold molecules in the viral replication complex. Viral replicase molecules distributed in a variety of aggregation states in different domains of the replication compartment (Figure 4) (Fernandez de Castro et al., 2014 and 2017).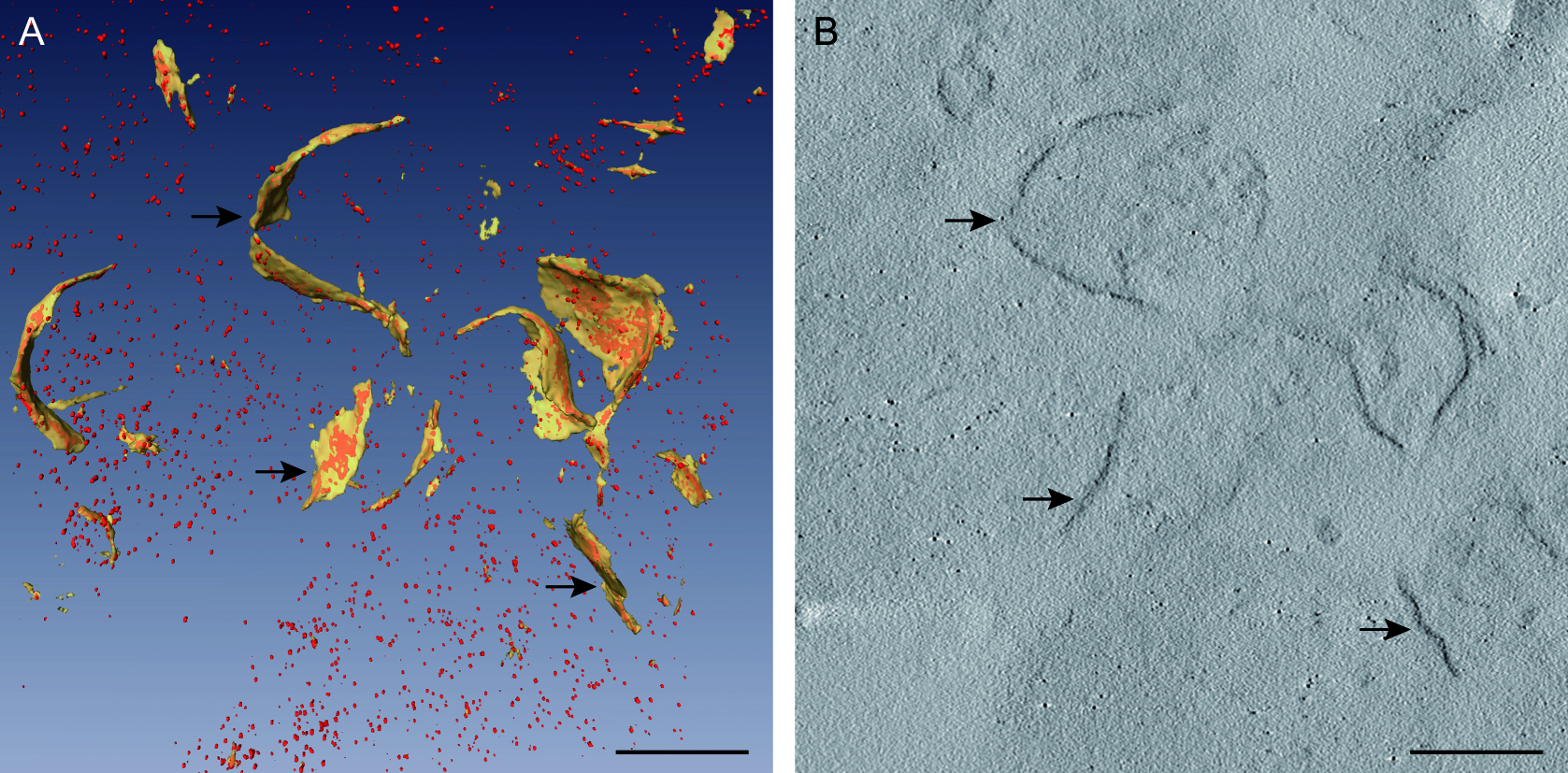
Figure 4. Electron tomography of the viral replication organelles. A. 3D model of the replication platform. Membranes are shown in yellow and p33–MT-gold molecules in red (arrows). B. Computational tomographic slice corresponding to the replication platform showed in A. Arrows point to aggregates of p33–MT-gold molecules inside membranes. Nano-clusters have an approximate diameter of 1 nm. Scale bars = 100 nm.
Notes
- Analyze the sequence of the protein of interest in order to identify the best site for inserting the MT tag. In addition, it is relevant to test if the MT-tagged protein activity and function are affected.
- Adjust the optimal conditions in your system for expression of the MT-tagged protein.
- Make a preliminary EM study to get familiar with the ultrastructure of the cell type. This will facilitate data interpretation.
- Check that the gold salts do not precipitate in the medium where cells are going to be treated.
- Incubate control cells without MT-tagged proteins with gold salts and visualize electron microscopy to verify lack of nonspecific background.
- Use acrylic resin (such as LR White or Lowicryl) in the absence of staining agents to visualize the MT-nanoclusters.
- Perform immunogold labeling with antibodies specific for the MT-tagged protein to confirm labeling specificity.
Recipes
- YPD
10 g yeast extract
20 g peptone
20 g dextrose (glucose)
Add dH2O to 1,000 ml and mix well
Autoclave to sterilize
For plates add 20 g of agar/L - YPG
10 g yeast extract
20 g peptone
20 g galactose
Add dH2O to 1,000 ml and mix well
Autoclave to sterilize
For plates add 20 g of agar/L - Transformation mix
0.36 ml 1 M lithium acetate
0.1 ml 10 mg/ml ssDNA
0.69 ml dH2O
2.4 ml 50%PEG MW3350
Add dH2O up to 10 ml - TSD reduction buffer
0.1 M Trizma® hemisulfate, pH 9.4
10 mM DTT (added just before use) - Spheroplast medium A
1x yeast nitrogen base
2% (w/v) glucose
1x amino acids
1 M sorbitol
20 mM Tris-HCl, pH 7.5
Store up to 4 weeks at room temperature - 4% paraformaldehyde (PFA) solution
- Dissolve 4 g of PFA in 90 ml of distilled water at 60 °C under a well ventilated hood
- Add drops of 1 N NaOH until the solution is transparent and without precipitates
- Then remove from heating and cool down on ice
- Add 10 ml of 10x PBS and fill up with distilled water to 100 ml
- Control the temperature and avoid heating the solution above 70 °C because PFA will break down at higher temperatures
- Filter the solution with 37 mm syringe filters
- Use the solution immediately or store for 1 day at 4 °C or at -20 °C for months
- Dissolve 4 g of PFA in 90 ml of distilled water at 60 °C under a well ventilated hood
- PHEM solution, pH 6.9
20 mM PIPES
50 mM HEPES
20 mM EGTA
4 mM MgCl2
Store for several months at 4 °C
Acknowledgments
We would like to express our gratitude to Drs José Jesús Fernández and Peter Nagy for expert advice and helpful discussions. This work was supported by grant BIO2015-68758-R (to C.R.) from the Spanish Ministry of Economy and Competitiveness (MINECO-AEI/FEDER, EU). The protocol is adapted from Fernandez de Castro et al. (2017). The authors declare no conflicts of interest.
References
- Barajas, D., Martin, I. F., Pogany, J., Risco, C. and Nagy, P. D. (2014a). Noncanonical role for the host Vps4 AAA+ ATPase ESCRT protein in the formation of Tomato bushy stunt virus replicase. PLoS Pathog 10(4): e1004087.
- Barajas, D., Xu, K., de Castro Martin, I. F., Sasvari, Z., Brandizzi, F., Risco, C. and Nagy, P. D. (2014b). Co-opted oxysterol-binding ORP and VAP proteins channel sterols to RNA virus replication sites via membrane contact sites. PLoS Pathog 10(10): e1004388.
- Bouchet-Marquis, C., Pagratis, M., Kirmse, R. and Hoenger, A. (2012). Metallothionein as a clonable high-density marker for cryo-electron microscopy. J Struct Biol 177(1): 119-127.
- Choi, H. S., Su, W. M., Morgan, J. M., Han, G. S., Xu, Z., Karanasios, E., Siniossoglou, S. and Carman, G. M. (2011). Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER602, THR723, AND SER744 as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J Biol Chem 286(2): 1486-1498.
- de Castro, I. F., Volonte, L. and Risco, C. (2013). Virus factories: biogenesis and structural design. Cell Microbiol 15(1): 24-34.
- de Castro Martin, I. F., Fournier, G., Sachse, M., Pizarro-Cerda, J., Risco, C., & Naffakh, N. (2017). Influenza virus genome reaches the plasma membrane via a modified endoplasmic reticulum and Rab11-dependent vesicles. Nat Commun 8(1): 1396.
- Delebecque, C. J., Lindner, A. B., Silver, P. A. and Aldaye, F. A. (2011). Organization of intracellular reactions with rationally designed RNA assemblies. Science 333(6041): 470-474.
- den Boon, J. A., Diaz, A. and Ahlquist, P. (2010). Cytoplasmic viral replication complexes. Cell Host Microbe 8(1): 77-85.
- Diestra, E., Fontana, J., Guichard, P., Marco, S. and Risco, C. (2009). Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol 165(3): 157-168.
- Fernandez de Castro, I., Fernandez, J. J., Barajas, D., Nagy, P. D. and Risco, C. (2017). Three-dimensional imaging of the intracellular assembly of a functional viral RNA replicase complex. J Cell Sci 130(1): 260-268.
- Fernandez de Castro, I., Sanz-Sanchez, L. and Risco, C. (2014). Metallothioneins for correlative light and electron microscopy. Methods Cell Biol 124: 55-70.
- Jonczyk, M., Pathak, K. B., Sharma, M. and Nagy, P. D. (2007). Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362(2): 320-330.
- McCartney, A. W., Greenwood, J. S., Fabian, M. R., White, K. A. and Mullen, R. T. (2005). Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17(12): 3513-3531.
- Mercogliano, C. P. and DeRosier, D. J. (2006). Gold nanocluster formation using metallothionein: mass spectrometry and electron microscopy. J Mol Biol 355(2): 211-223.
- Mercogliano, C. P. and DeRosier, D. J. (2007). Concatenated metallothionein as a clonable gold label for electron microscopy. J Struct Biol 160(1): 70-82.
- Miller, S. and Krijnse-Locker, J. (2008). Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol 6(5): 363-374.
- Nagy, P. D. (2008). Yeast as a model host to explore plant virus-host interactions. Annu Rev Phytopathol 46: 217-242.
- Nagy, P. D. (2016). Tombusvirus-host interactions: Co-opted evolutionarily conserved host factors take center court. Annu Rev Virol 3(1): 491-515.
- Nagy, P. D. and Pogany, J. (2011). The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10(2): 137-149.
- Risco, C., Sanmartin-Conesa, E., Tzeng, W. P., Frey, T. K., Seybold, V. and de Groot, R. J. (2012). Specific, sensitive, high-resolution detection of protein molecules in eukaryotic cells using metal-tagging transmission electron microscopy. Structure 20(5): 759-766.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Fernandez de Castro, I. and Risco, C. (2018). Metal-tagging Transmission Electron Microscopy for Localisation of Tombusvirus Replication Compartments in Yeast. Bio-protocol 8(8): e2822. DOI: 10.21769/BioProtoc.2822.
Category
Microbiology > Microbe-host interactions > Virus
Cell Biology > Cell imaging > Electron microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link