- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of Polyhydroxybutyrate (PHB) Content in Ralstonia eutropha Using Gas Chromatography and Nile Red Staining
(*contributed equally to this work) Published: Vol 8, Iss 5, Mar 5, 2018 DOI: 10.21769/BioProtoc.2748 Views: 17987
Reviewed by: Dennis NürnbergBenoit ChassaingAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1764 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1808 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1779 Views
Abstract
Ralstonia eutropha H16 produces and mobilizes (re-utilizes) intracellular polyhydroxybutyrate (PHB) granules during growth. This protocol describes the visualization of intracellular Nile red stained PHB granules and the quantification of PHB by gas chromatography. Our first method describes how to analyze PHB granules by fluorescence microscopy qualitatively. Our second approach enables the conversion of PHB to volatile hydroxycarboxylic acid methyl esters by acidic methanolysis and their quantification by gas chromatography. Through this method, it is possible to obtain an absolute quantification of PHB, e.g., per cell dry weight.
Keywords: Polyhydroxybutyrate (PHB)Background
Polyhydroxyalkanoates (PHA), especially polyhydroxybutyrate (PHB), are energy and carbon storage compounds in many prokaryotic species, ensuring bacterial survival under stress conditions (Anderson and Dawes, 1990; Pötter and Steinbüchel, 2006; Jendrossek and Pfeiffer, 2014; Bresan et al., 2016). An industrial application of these biopolymers is the production of biodegradable plastic (Chen, 2009; Riedel et al., 2015) and the research on potential medicinal components (Wu, 2009; Zonari et al., 2015; Pacheco et al., 2015; Giretova et al., 2016). Ralstonia eutropha H16, a Gram-negative facultative chemolithoautotrophic β-proteobacterium, is a model organism for PHB accumulation as it can accumulate up to 80% of its cell dry weight of PHB. Within the cells, PHB forms granules or so-called carbonosomes covered with different surface proteins (Jendrossek and Pfeiffer, 2014; Bresan et al., 2016). PHB is synthesized from its parent substance acetyl-CoA in a 3-step reaction. The first step is a condensation reaction of two acetyl-CoA molecules by the acetyl-CoA-acetyltransferase PhaA. Acetoacetyl-CoA is then reduced to (R)-3-hydroxybutyryl-CoA by the acetoacetyl-CoA-reductase PhaB. The last step includes an essential non-redundant reaction: the polymerization of (R)-3-hydroxybutyryl-CoA to PHB by the PHB synthase called PhaC (Figure 1).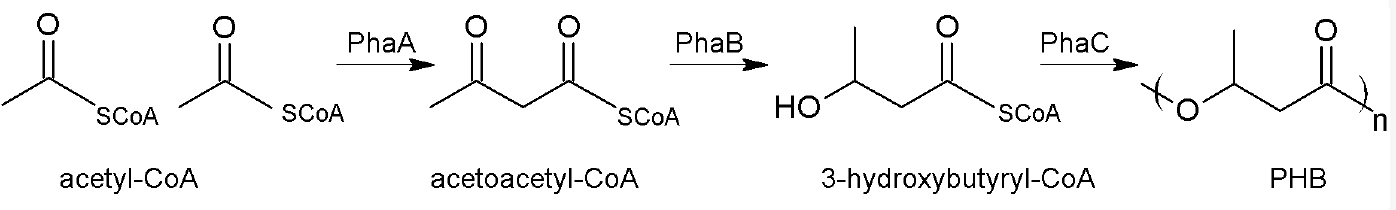
Figure 1. Biosynthesis of PHB
A fast and easy way to detect intracellular PHB is a microscopy approach using Nile red staining. Nile red (also known as Nile blue oxazone) is a lipophilic fluorescent dye used to visualize hydrophobic cell structures such as membranes or lipid-like inclusions (PHB, triacyl-glycerides) (Spiekermann et al., 1999). Nile red binds to PHB granules and can easily be detected by fluorescence microscopy. Its colors (i.e., fluorescent emission wave lengths) vary from dark red (for binding to polar membrane lipids) to an intense yellow-gold emission (for binding to neutral lipids in intracellular storages). The emission (> 590 nm) and excitation (560 nm) wavelengths characteristic of the Nile red hydrophobic compound adducts also depend on solvent polarity (Spiekermann et al., 1999); in most polar solvents Nile red shows no or only little fluorescence.
Gas chromatography (GC) can be used to quantify PHB and to determine its monomeric composition. PHB decomposes at temperatures below its boiling point. Therefore, PHB must be converted into products that are stable and volatile at the temperature of the GC-column. This is achieved by conversion of PHB into volatile hydroxycarboxylic acid methyl esters, hereafter, methyl esters (Figure 2) (Brandl et al., 1988). The methyl esters interact specifically with the solid phase thereby allowing a separation of different hydroxyalkanoate methyl esters in case co-polyesters of different hydroxyalkanoates have to be analyzed. Measuring the time point of appearance and the area under the resulting compound peak of the detector signals in the chromatogram enable its quantitative and qualitative determination.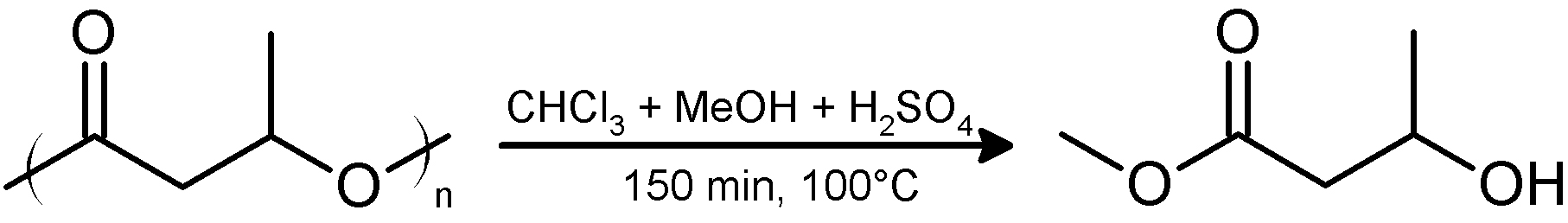
Figure 2. Acidic methanolysis of PHA
Materials and Reagents
- Microscope slides (e.g., Carl Roth, catalog number: H868.1 )
- Cover slips (e.g., Carl Roth, catalog number: H873.2 )
- 2 ml reaction tubes (e.g., SARSTEDT, catalog number: 72.695.500 )
- 50 ml Falcon tubes (e.g., SARSTEDT, catalog number: 62.559.001 )
- 6 ml culture tubes with screw-cap with chloroform resistant PTFE seal (e.g., DWK Life Sciences, DURAN, catalog number: 26 135 11 5 )
- GC glass vial (e.g., Brown, catalog number: 155710 )
- 50 ml Omnifix® Syringes (e.g., B.Braun Medical, catalog number: 4591281 )
- Sterile filter Filtropur S 0.2 (e.g., SARSTEDT, catalog number: 83.1826.001 )
- Scalpel blade (e.g., Gebrüder Martin, KLS Martin, catalog number: 10-155-24-04 )
- Pipette tips 1,000 μl (e.g., SARSTEDT, catalog number: 70.762.010 )
- Pipette tips 200 μl (e.g., SARSTEDT, catalog number: 70.760.002 )
- Pipette tips 10 μl (e.g., VWR, catalog number: 53509-070 )
- 1.5 ml tubes (e.g., SARSTEDT, catalog number: 72.690.001 )
- 0.3 ml limited volume inserts (e.g., Brown, catalog number: 155650 )
- Septa (e.g., Brown, catalog number: 155615 )
- Organisms
- Ralstonia eutropha H16 (alternative strain designations: Hydrogenomonas eutropha H16, Alcaligenes eutrophus H16, Wautersia eutropha H16, Cupriavidus necator H16). DSM 428 (Deutsche Sammlung für Mikroorganismen and Zellkulturen GmbH, https://www.dsmz.de/). Wild type strain produces PHB and related short-chain-length PHA
- Ralstonia eutropha H16-PHB-4 (DSM 541), PHB negative mutant of strain H16 because of mutation G320A in the PHB synthase (phaC) gene (Raberg et al., 2014)
- Ralstonia eutropha H16 (alternative strain designations: Hydrogenomonas eutropha H16, Alcaligenes eutrophus H16, Wautersia eutropha H16, Cupriavidus necator H16). DSM 428 (Deutsche Sammlung für Mikroorganismen and Zellkulturen GmbH, https://www.dsmz.de/). Wild type strain produces PHB and related short-chain-length PHA
- PHB (e.g., Sigma-Aldrich, catalog number: 363502 )
- Agarose standard (e.g., Carl Roth, catalog number: 3810.4 )
- Nitrogen gas (e.g., Air Liquide, ALPHAGAZTM 1 Stickstoff, catalog number: P0271L50R2A001 )
- Helium gas (e.g., Air Liquide, ALPHAGAZTM 1 Helium, AIR LIQUIDE Deutschland, catalog number: P0251L50R2A001 )
- Synthetic air (e.g., Air Liquide, ALPHAGAZTM 1 Luft, AIR LIQUIDE Deutschland, catalog number: P0291L50R2A001 )
- Octane (e.g., Sigma-Aldrich, catalog number: 74821 )
- Nile red (e.g., Sigma-Aldrich, catalog number: N3013 )
- DMSO (e.g., Carl-Roth, catalog number: 7029.2 )
- Trichloromethane/Chloroform (e.g., Carl Roth, catalog number: 6340.2 )
- Methanol for GC (e.g., VWR, catalog number: 20864.320 )
- Methyl benzoate (e.g., Sigma-Aldrich, catalog number: M29908 )
- Sulphuric acid 96% (e.g., Carl Roth, catalog number: 4623.1 )
- Fructose
- Nutrient broth (e.g., BD, DifcoTM, catalog number: 231000 )
- Na2HPO4·12H2O
- KH2PO4
- NH4Cl
- MgSO4·7H2O
- CaCl2·7H2O
- Ferric ammonium citrate
- ZnSO4
- MnCl2·4H2O
- H3BO3
- CoCl2·6H2O
- CuCl2·2H2O
- NiCl2·6H2O
- NaMoO4·2H2O
- D-Gluconic acid sodium salt (e.g., Sigma-Aldrich, catalog number: G9005 )
- NB medium (see Recipes)
- Mineral salts medium (see Recipes)
- D-Gluconic acid sodium salt solution (20% stock solution) (see Recipes)
- Nile red solution (see Recipes)
Equipment
- 100 ml Erlenmeyer flasks (e.g., DWK Life Sciences, DURAN, catalog number: 21 216 24 )
- 500 ml Erlenmeyer flasks (e.g., DWK Life Sciences, DURAN, catalog number: 21 216 44 )
- 3 L Erlenmeyer flasks (e.g., DWK Life Sciences, DURAN, catalog number: 21 216 68 )
- Incubation shaker (e.g., INFORS HT)
- Pipettes (e.g., Thermo Scientific)
- Spatula
- Centrifuge (e.g., Eppendorf, model: 5417 C )
- Freeze-dryer (e.g., Christ, model: Alpha 1-2 LDplus )
- Rotary vane pumps (e.g., Pfeiffer Vacuum, model: DUO 5 M )
- Analytical balance (e.g., Sartorius, model: A 200 S )
- Fume hood
- Oil bath (e.g., Memmert)
- Gas chromatograph (e.g., Agilent Technologies, model: Agilent 7890A ; flame ionization detector (FID))
- Gastight syringe for GC (e.g., VWR, catalog number: 5490572)
Manufacturer: Hamilton, model: 1701 SN CTC . - CTC automated sample injector (e.g., Agilent Technologies, catalog number: G6501-CTC )
- GC column DB-WAX (e.g., Agilent Technologies, catalog number: 122-7032 )
- Fluorescence microscope with a Plan Apo objective (100x/1.4 oil) (e.g., Nikon Instruments, model: Eclipse Ti-E )
- Nile red-Filter (Excitation: 562/40 nm/Emission: 594 (long pass), e.g., AHF Analysentechnik AG, Tübingen, Germany, www.ahf.de/)
- Liner 4 mm ID LPD (e.g., Agilent Technologies, catalog number: 5183-4647 )
- Freezer
- Sterile bench (e.g., HERA safe)
- Refrigerated Falcon centrifuge (e.g., Sigma Zentrifugen, model: 4K15 )
- Vortex
- Laboratory glass bottles (e.g., DWK Life Sciences, DURAN, catalog number: 21 801 54 5 )
- Autoclave
Software
- GC ChemStation Rev. B.04.01 SP1, Agilent
- Excel, Microsoft, Redmont, USA
- Nikon imaging software
- ImageJ Fiji vl.50c
Procedure
Note: Ensure that all safety instructions for the handling of hazardous compounds and for waste management are properly considered; since these may vary in different countries the following protocol does not provide any instruction on these issues.
Part I. Nile red staining
See Figure 3 for the outline of Nile red staining procedure.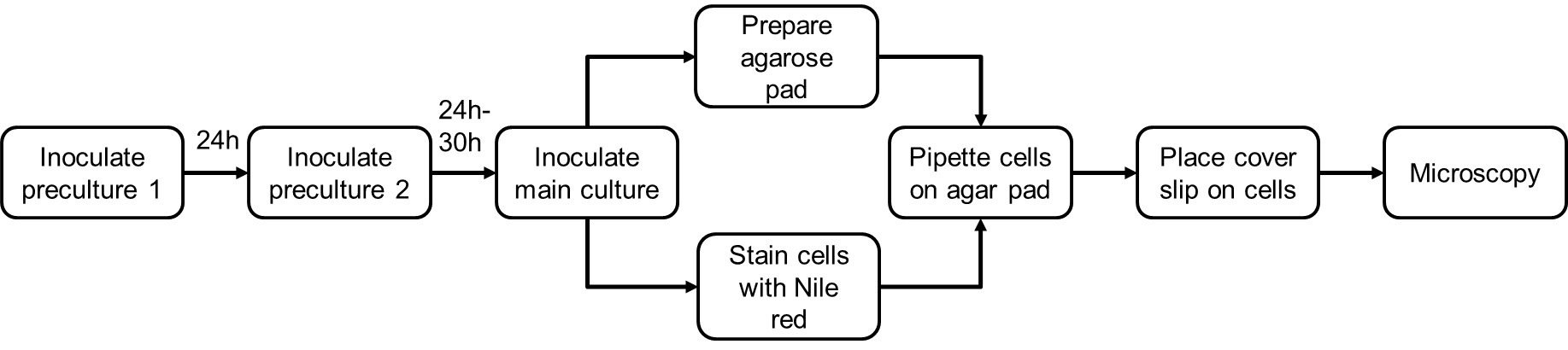
Figure 3. Flow chart the Nile red staining
Note: Nile red staining can be performed with cells taken either from cultures for gas chromatography analysis or from independent cultures prepared for microscopy experiments.
- Preparations of cells
- Inoculate a first seed culture of 10 ml NB medium (or of a culture medium that allows good growth of the species to investigate) with a single colony of R. eutropha H16 cells in a 100 ml flask and incubate the flask for 24 h on a shaker at 150 rpm and 30 °C.
- Inoculate a second seed culture of 9 ml NB medium with 1 ml of the first seed culture (1:10 dilution) in a 100 ml flask. Incubate the cells for 24-30 h on a shaker at 150 rpm and 30 °C. The procedure of two subsequent seed cultures on NB medium provides R. eutropha cells that are in the stationary growth phase as revealed by the presence of mainly short-rod-shaped or almost coccoid cells. Most of the cells (> 95%) have mobilized any previously accumulated PHB and the cells appear to be ‘empty’ after Nile red staining.
- Inoculate the main culture by transferring 1 ml of the second pre-culture to 9 ml of fresh NB medium (in a 100 ml flask) supplemented with 0.2% D-gluconic acid sodium salt (100 μl of 20% stock solution) and incubate the cells on a shaker at 150 rpm and 30 °C. Gluconate increases the C to N ratio of the medium and promotes accumulation of PHB. If different species are investigated other C-sources that a metabolized via acetyl-coenzyme A (precursor of PHB) may be added to promote the formation of PHB, e.g., acetate or glucose.
- Inoculate a first seed culture of 10 ml NB medium (or of a culture medium that allows good growth of the species to investigate) with a single colony of R. eutropha H16 cells in a 100 ml flask and incubate the flask for 24 h on a shaker at 150 rpm and 30 °C.
- Preparation of microscope slides
- Label the microscope slides.
- Prepare agarose pads by pipetting 100 µl of hot (~60 °C) agarose solution (1% [w/v] in H2O) on the slide and immediately place the cover slip on the agarose (Figure 4).

Figure 4. Preparation of an agarose pad for microscopy - Let it solidify (≥ 2 min) and carefully remove the cover slip using a blade. Having removed the cover slip the sample should be applied within the next two minutes. Otherwise, the agar surface will become dry.
- Label the microscope slides.
- Nile red staining of cells
- Take samples every 2 h after inoculation of the main culture during the accumulation and mobilization phase of PHB. Remaining cells of the second pre-culture can be used as a negative control and represent the time point ‘0’ (T0). These cells (> 95%) should have no accumulated PHB. Alternatively, cells of R. eutropha PHB-4 can be used as a negative control for PHB granule accumulation. Due to a mutation in the PHB synthase gene cells of R. eutropha PHB-4 are unable to synthesize storage PHB (Raberg et al., 2014).
- Harvest 1 ml of culture by centrifugation (60 sec, 13,000 x g), discard the supernatant and resuspend the pellet in the remaining ~30-50 µl medium that congregates from the tube walls at the bottom within ~1 min.
- Add 4 µl of cells to 1 µl of Nile red (working solution: 10 µg/ml in DMSO, light sensitive, stable for several months at 4 °C) in a reaction tube.
- Drop 1 or 2 µl of the stained cell suspension on the agarose pad, let it dry for a few seconds and carefully place the cover slip on the agarose.
- Take samples every 2 h after inoculation of the main culture during the accumulation and mobilization phase of PHB. Remaining cells of the second pre-culture can be used as a negative control and represent the time point ‘0’ (T0). These cells (> 95%) should have no accumulated PHB. Alternatively, cells of R. eutropha PHB-4 can be used as a negative control for PHB granule accumulation. Due to a mutation in the PHB synthase gene cells of R. eutropha PHB-4 are unable to synthesize storage PHB (Raberg et al., 2014).
- Microscopy analysis
- Image the samples using an appropriate fluorescence filter for Nile red (excitation: 562/40 nm, emission 594 (long pass), AHF Analysentechnik AG, Tübingen, Germany, www.ahf.de). Image the cells also under bright field.
- Analyze the pictures with the Nikon imaging software or ImageJ Fiji vl.50c.
- Image the samples using an appropriate fluorescence filter for Nile red (excitation: 562/40 nm, emission 594 (long pass), AHF Analysentechnik AG, Tübingen, Germany, www.ahf.de). Image the cells also under bright field.
Part II. Determination of PHB content using gas chromatography
See Figure 5 for the outline of the procedure to determine the PHB content using gas chromatography.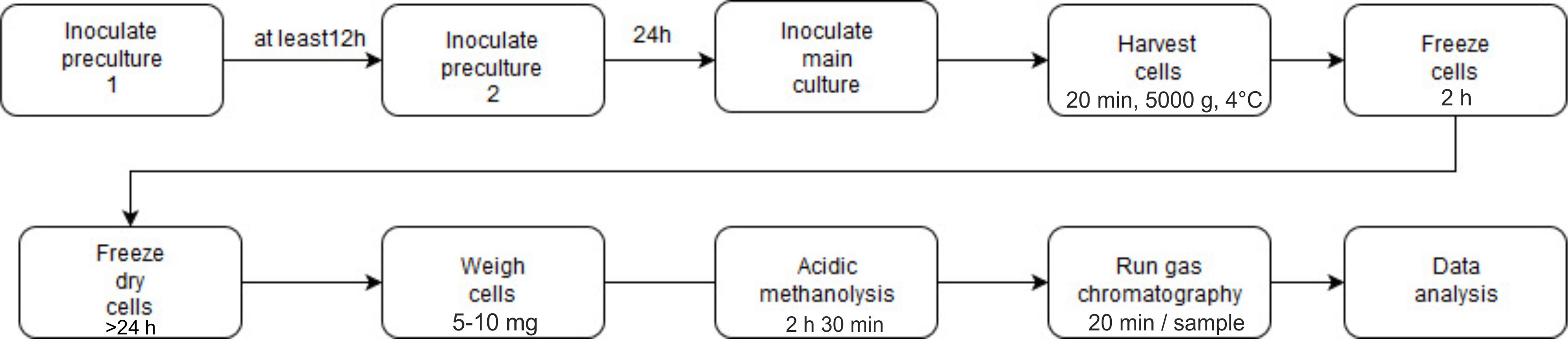
Figure 5. Flow chart of the determination of PHB content using gas chromatography
- Cultivation and harvesting of cells
- Three independent colonies and cultures should be used for biological triplicates.
- Calculate the appropriate amount of culture volume depending on how many samples (data points) will be taken. 100 ml of culture is necessary for a data point when the culture has an OD600 ≤ 0.6. 50 ml of culture is necessary for a data point when the culture has an OD600 ≥ 0.6. We usually take a sample every 4 h over a period of 48 h.
- Inoculate the first seed cultures with 10 ml NB medium, the appropriate antibiotics if necessary and a colony of R. eutropha H16 cells and incubate it overnight in a shaker at 30 °C up to 24 h.
- Inoculate the second seed cultures with a dilution of 1:10 in NB medium and the appropriate antibiotics if necessary. Incubate it for at least 24-30 h in a shaker at 30 °C to get rid of all previously accumulated PHB. The culture should have a volume of at least 50 ml to have enough cells to harvest the T0 time point from the seed culture and to inoculate the main culture.
- Inoculate the main cultures with a dilution of 1:20 in NB medium, 0.2% sodium gluconate and the appropriate antibiotics if necessary and incubate it in a shaker at 30 °C.
- Take 50-100 ml samples every 4 h during the accumulation and mobilization phase of PHB. T0 is represented by the second pre culture. The volume of the main culture should be 50 ml higher than the sum of the volumes harvested for all data points.
- Harvest 50-100 ml of culture in 50 ml Falcon tubes by centrifugation for 20 min at 5,000 x g and 4 °C.
- Discard the supernatant and freeze the pellet for at least 2 h at -20 °C.
- Three independent colonies and cultures should be used for biological triplicates.
- Lyophilization and weighing of samples
- In order to freeze-dry the cell pellets, slightly open the lids of the Falcon tubes or use lids with a prepared hole.
- Place the Falcon tubes in the freeze-dryer and start the drying process by switching on the (rotary) vacuum pump. Freeze-dry the samples for at least 24 h. Lyophilization of the samples is finished when the cell pellets are dry. The absence of any residual water/moisture is important.
- Disrupt all cell pellets into small clumps using a spatula.
- Weigh approximately 10 mg of the freeze-dried cell pellets on an analytical balance into screw-capped culture tubes with chloroform resistant PTFE seal. If the size of the pellet is limited, it is possible to use fewer amounts of freeze-dried cells but the (mass) weight should always be between 5-10 mg.
Note: It is important to determine the exact cell mass (weight) for later calculations. - In order to generate a PHB calibration graph, weigh 2 mg, 4 mg, 6 mg and 8 mg of pure PHB into culture tubes with screw-caps, respectively.
- In order to freeze-dry the cell pellets, slightly open the lids of the Falcon tubes or use lids with a prepared hole.
- Acidic methanolysis and preparation of GC samples (Figure 6)
Note: All steps should be performed in a fume hood.- Add 1 ml of chloroform to the culture tubes.
- Add 1 ml of methanol supplemented with 15% (v/v) H2SO4 (150 μl H2SO4 + 850 μl methanol per sample) to the special culture tubes containing the weighted dried cells. Close the tubes tightly and vortex for three seconds. All pellet clumps should be in the solvent mixture.
- Incubate the tubes for 2 h 30 min at 100 °C in a thermostat-equipped oil bath. Follow the local safety instructions as the applied temperature is above the boiling point of methanol and chloroform. Intact sealing and the absence of any fissures in the glass tubes are essential.
- Cool down the samples on ice for 5 min.
- Add 1 ml of deionized water and 1 ml of chloroform containing 0.2% (v/v) methyl benzoate (2 μl methyl benzoate + 998 μl chloroform per sample) as an internal standard.
Note: Use the same chloroform-methyl benzoate mixture for all samples to avoid dilution errors. - Close the tubes tightly and vortex vigorously for 30 sec.
- Let the tubes stand for a minute to allow phase separation.
- Pipette 150 µl of the organic (bottom) phase into 0.3 ml limited volume inserts in GC glass vials.
- Close the screw cap of the GC vials.
- Prepare a GC vial with 150 µl pure chloroform as equilibration solvent for the column.
- Pipette 1 ml of pure chloroform and 1 ml of the chloroform methyl benzoate mixture into a culture tube to prepare an external standard. Transfer 150 µl of the mixture into a GC vial.
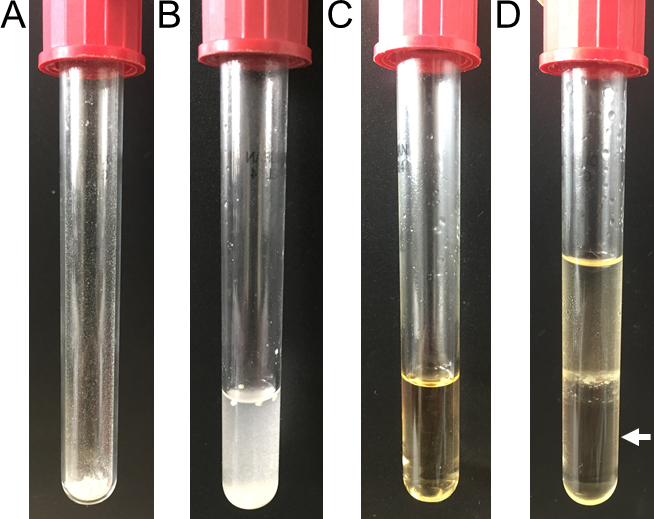
Figure 6. Images of the acidic methanolysis steps of PHB. Weighted cells (A), complete methanolysis sample before oil bath (B), sample after oil bath (C) and phase-separated sample with the bottom organic phase (white arrow, D).
- Add 1 ml of chloroform to the culture tubes.
- GC measurement
- Place all GC vials in a GC tray.
- Refill all glass vials which are used to rinse the syringe three times between different samples with octane.
- Empty all waste vials.
- Set up the GC apparatus and the CTC automated sample injector according to the instructions supplied by the manufacturer and install a gastight syringe as well as the appropriate liner.
- Use a DB-WAX column as stationary phase and the inert gas helium as mobile phase for the separation of the methyl esters.
- Program an injection volume of 1 µl, a split mode with a split ratio of 1:8 and an injection temperature of 250 °C.
- Program a flow rate of 0.7 ml/min.
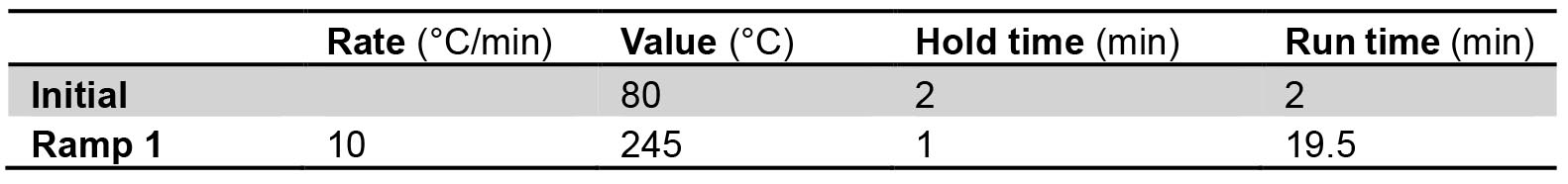
- Program the following temperature gradient for the separation of the methyl esters:
- Detection is performed with a flame ionization detector (FID) at 275 °C.
- Place all GC vials in a GC tray.
Data analysis
Part I. Nile red staining–Image analysis
Analyze the taken pictures using the Nikon imaging software or ImageJ Fiji vl.5 as shown in Figure 7.
Figure 7. Fluorescence micrographs of cells with and without PHB. From left to right: bright field, Nile red, merged channels of bright field and Nile red. Upper panel: cells with PHB granules, Lower panel: cells without PHB granules.
Note: That cells can have ‘dark’ inclusions (e.g., most upper cell in lower panel) that are not stained with Nile red. Such inclusions might represent other inclusion bodies/structures (e.g., polyphosphate granules).
Part II. Determination of PHB content using gas chromatography
- Double click on the sample name of interest to open the chromatogram.
- The 3-hydroxybutyryl methyl ester (3HBME) should elute at 7.0-7.1 min. The internal standard (methyl benzoate) should elute at 8.7-8.8 min. The exact retention times are apparatus- and condition-specific and might vary. If a copolymer of 3HB and 3-hydroxyvalerate (3HV) is accumulated by the cells the retention times of 3HV methylester (3HVME) is around 7.9-8.0 min. The same protocol can also be applied for the analysis and detection of medium-chain-length PHAs. The methyl esters of the 3-hydroxyalkanoic acids with 6, 8, 10 and 12 C-atoms elute at ~9.0 min (C6), ~11.4 min (C8), ~13.5 min (C10) and ~15.5 min (C12), respectively (not shown).
- Click on the integration button (Red square in Figure 8).
- In the event that the peak is too small to be automatically integrated, it can be manually integrated by pressing the manual integration button (Green box in Figure 8) and drawing the pink line under the peak with the mouse cursor.
- The values of the area under the curve will be listed in a table under the chromatogram (Orange box in Figure 8).
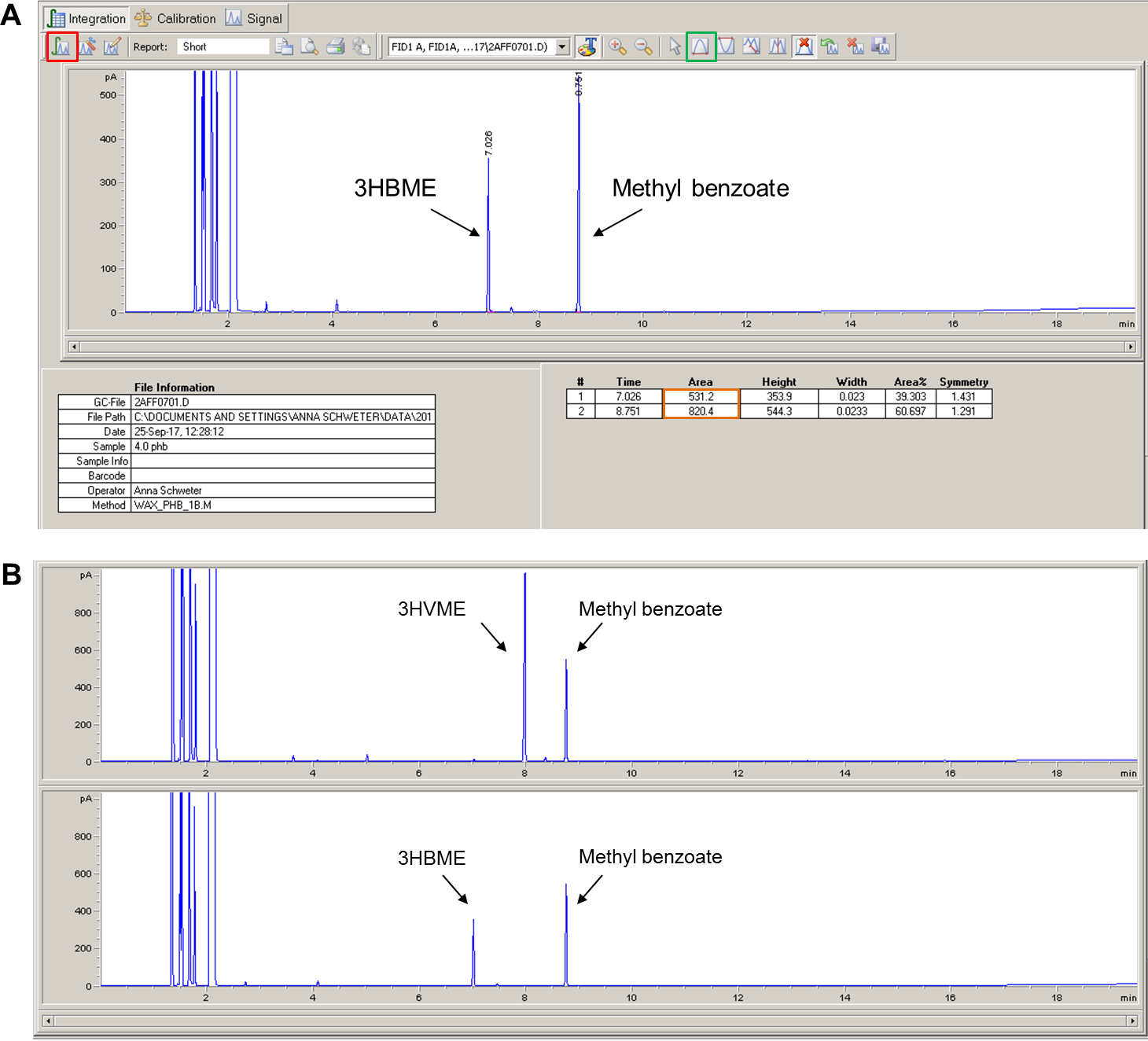
Figure 8. Screenshot of a chromatogram and results tables (A) and representative chromatograms for 3HVME and 3HBME (B) - Copy the values of the area under the 3-hydroxybutyryl methyl ester (3-HBME) peak and the methyl benzoate peak to an excel table for all samples of the PHB standard curve and for all samples to be analyzed.
- Calculate the normalized area of the 3-HBME peaks using the formula:

- Create a standard curve by plotting the normalized areas of the PHB standard samples on the y-axis against the weighted amounts of PHB on the x-axis (Figure 9) and determine the equation of the function (y = a*x + b).
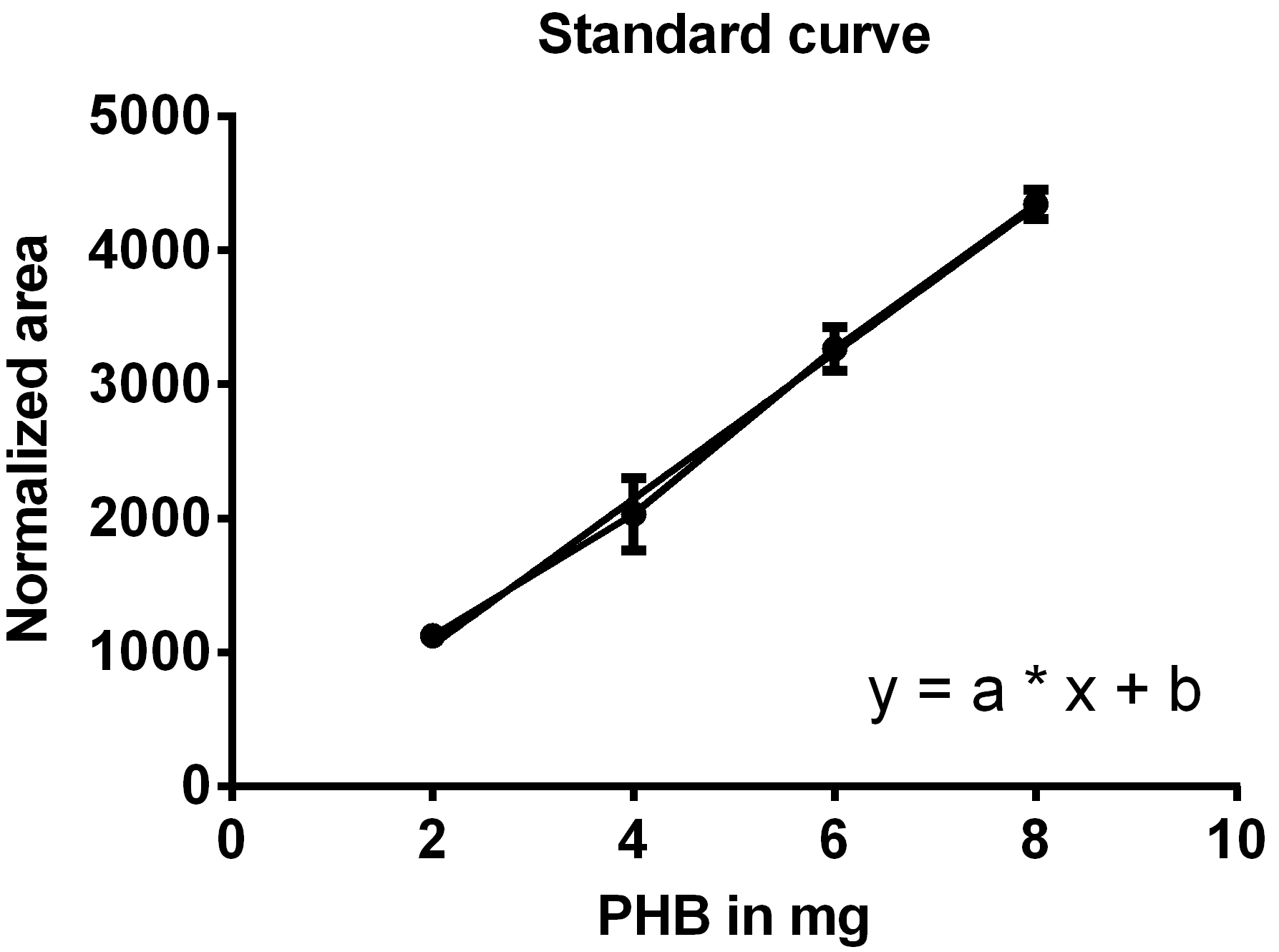
Figure 9. Standard curve for the determination of PHB weight. One line connects the data points, the second line is the trend line used to generate the equation of the line. - Calculate the weight of PHB in your sample using the formula:
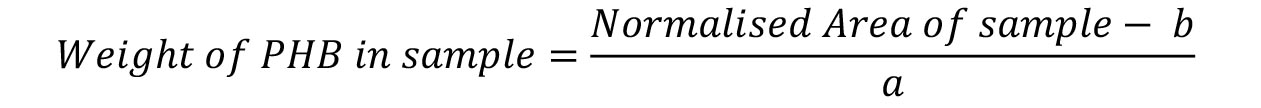
- Calculate the percentage of PHB per cell dry weight using the formula:
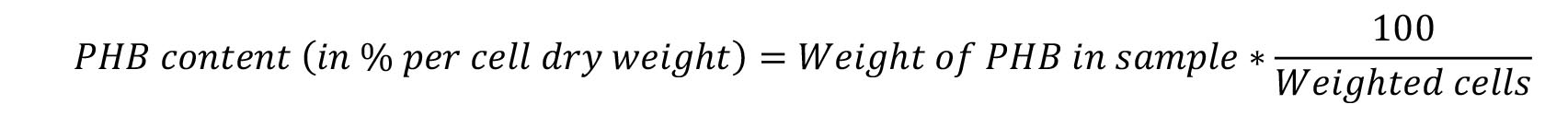
- Represent the PHB content in the graph with the program of your choice (Figure 10).
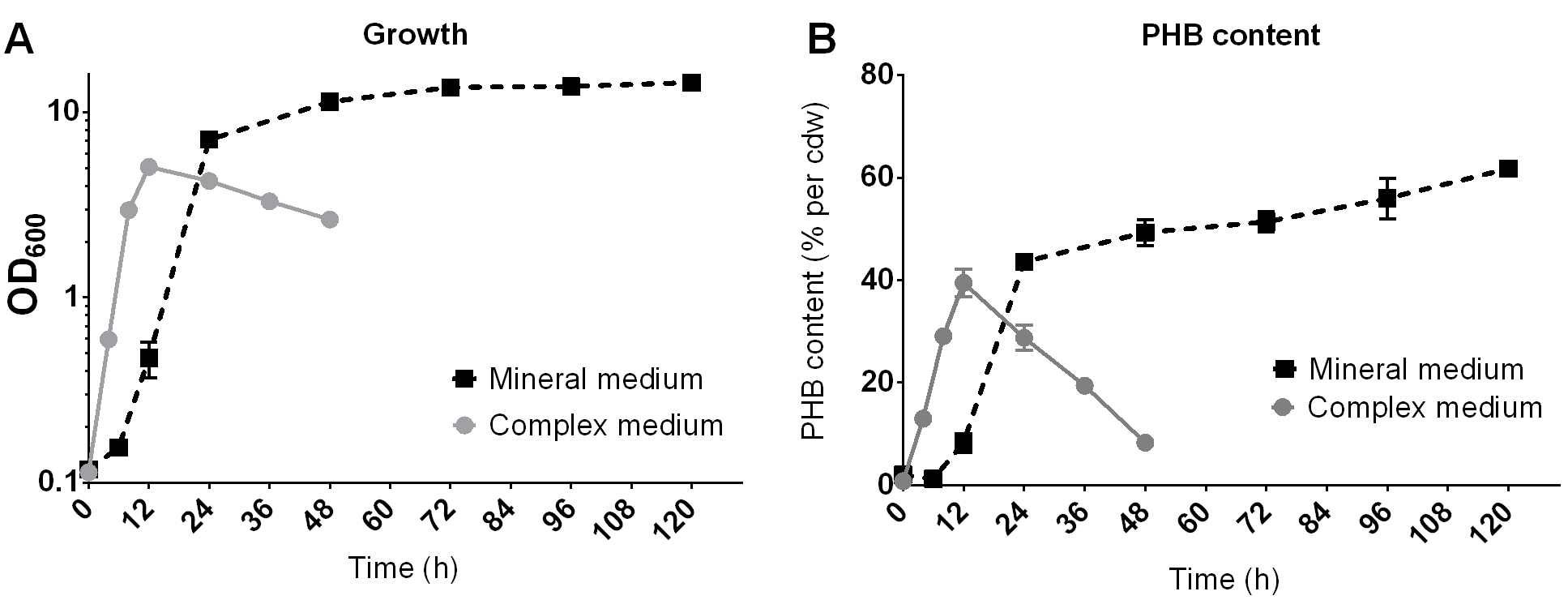
Figure 10. Example of the growth (A) and PHB production (B) of R. eutropha H16 on NB-medium with 0.2% Na-gluconate (complex medium) or on mineral salts medium with 2% fructose
Recipes
- NB medium
- Weigh 8 g of NB powder, fill it into a 1,000 ml glass bottle and add 1,000 ml of deionized water. Mix it and autoclave it at 121 °C for 20 min
- Store the NB medium at room temperature
- Weigh 8 g of NB powder, fill it into a 1,000 ml glass bottle and add 1,000 ml of deionized water. Mix it and autoclave it at 121 °C for 20 min
- Mineral salts medium
Any mineral salt medium that is suited for growth of the strain of interest can be used. For Ralstonia eutropha H16, we use the following medium. For preparation of 1,000 ml medium:- Dissolve 9 g Na2HPO4·12H2O, 1.5 g KH2PO4 and 1 g NH4Cl in ≈ 950 ml deionized water using a magnetic stirrer
- Add 1 ml each of the following 1,000x stock solutions:
Mg-solution (200 g/L MgSO4·7H2O)
Ca-solution (20 g/L CaCl2·7H2O)
Fe-solution (1.2 g/L ferric ammonium citrate)
Note: It is important that a magnetic stirrer is operating during the addition of Mg-, Ca-, and Fe- solutions. Otherwise white precipitates might form. - Finally, add 100 µl of a trace element solution
Values per liter trace element stock solution: 1 g ZnSO4, 0.3 g MnCl2·4H2O, 3 g H3BO3, 2 g CoCl2·6H2O, 0.1 g CuCl2·2H2O, 0.2 g NiCl2·6H2O, 0.3 g NaMoO4·2H2O and fill up to the end volume of 1,000 ml with deionized water - The pH of the mineral salts medium should be at pH 7.0 ± 0.1.
- Sterilize the medium at 121 °C for 20 min
- Minor amounts of precipitates will form upon sterilization that will partially re-dissolve upon cooling and stirring. Remaining small amounts of precipitates can be tolerated.
- Add the desired amount of carbon source before inoculation with the strain of interest (e.g., with sodium-gluconate stock solution up to 2% in case that high amounts of PHB should accumulate).
- Optionally, the amount of nitrogen source (NH4Cl) can be reduced to 0.5 g/L. This will lead to a higher C to N ratio and cause a reduction of the yield in cell biomass but will increase the % accumulated PHB per g cellular dry weight to ≈ 80%.
- Dissolve 9 g Na2HPO4·12H2O, 1.5 g KH2PO4 and 1 g NH4Cl in ≈ 950 ml deionized water using a magnetic stirrer
- D-Gluconic acid sodium salt solution (20% stock solution)
- Prepare a 20% wt/vol of D-gluconic acid sodium salt stock solution by dissolving 20 g sodium gluconate in 100 ml of deionized water
- Sterilize the solution by filtration through a sterile filter (0.2 µm) or by heat-sterilization (20 min 121 °C)
- The stock solution is stable at room temperature
- Prepare a 20% wt/vol of D-gluconic acid sodium salt stock solution by dissolving 20 g sodium gluconate in 100 ml of deionized water
- Nile red solution
- Prepare a Nile red stock solution of 1 mg/ml and dissolve it in DMSO
- Dilute the stock solution with DMSO to obtain a working solution of 10 µg/ml
- The Nile red solutions can be stored at 4 °C for several months
- Protect the solutions from light
- Prepare a Nile red stock solution of 1 mg/ml and dissolve it in DMSO
Acknowledgments
We thank Deutsche Forschungsgemeinschaft (DFG Je152/17-1) for funding. The authors declare no conflicts of interests.
References
- Anderson, A. J. and Dawes, E. A. (1990). Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54(4): 450-472.
- Brandl, H., Gross, R. A. and Lenz, R. W. (1988). Pseudomonas oleovorans, as a source of poly(beta-hydroxyalkanoates) for potential applications. Appl Env Microbiol 54:1977-1982.
- Bresan, S., Sznajder, A., Hauf, W., Forchhammer, K., Pfeiffer, D. and Jendrossek, D. (2016). Polyhydroxyalkanoate (PHA) granules have no phospholipids. Sci Rep 6: 26612.
- Chen, G. Q. (2009). A microbial polyhydroxyalkanoates (PHA) based bio- and materials industry. Chem Soc Rev 38:2434-2446.
- Giretova, M., Medvecky, L., Stulajterova, R., Sopcak, T., Briancin, J. and Tatarkova, M. (2016). Effect of enzymatic degradation of chitosan in polyhydroxybutyrate/chitosan/calcium phosphate composites on in vitro osteoblast response. J Mater Sci Mater Med 27:181.
- Jendrossek, D. and Pfeiffer, D. (2014). New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol 16(8): 2357-2373.
- Pacheco, D. P., Amaral, M. H., Reis, R. L., Marques, A. P. and Correlo, V. M. (2015). Development of an injectable PHBV microparticles-GG hydrogel hybrid system for regenerative medicine. Int J Pharm 478:398-408.
- Raberg, M., Voigt, B., Hecker, M. and Steinbüchel, A. (2014). A closer look on the polyhydroxybutyrate- (PHB-) negative phenotype of Ralstonia eutropha PHB-4. PLoS One 9:e95907.
- Riedel, S. L, Jahns, S., Koenig, S., Bock, M. C. E., Brigham, C. J., Bader, J. and Stahl, U. (2015). Polyhydroxyalkanoates production with Ralstonia eutropha from low quality waste animal fats. J Biotechnol 214:119-127.
- Spiekermann, P., Rehm, B. H., Kalscheuer, R., Baumeister, D. and Steinbüchel, A. (1999). A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch Microbiol 171:73-80.
- Wu, Q., Wang, Y. and Chen, G. Q. (2009). Medical application of microbial biopolyesters polyhydroxyalkanoates. Artif Cells Blood Substitutes Immobil Biotechnol 37:1-12.
- Zonari, A., Martins, T. M. M., Paula, A. C. C., Boeloni, J. N., Novikoff, S., Marques, A. P., Correlo, V. M., Reis, R. L. and Goes, A. M. (2015). Polyhydroxybutyrate-co-hydroxyvalerate structures loaded with adipose stem cells promote skin healing with reduced scarring. Acta Biomater 17:170-181.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Juengert, J. R., Bresan, S. and Jendrossek, D. (2018). Determination of Polyhydroxybutyrate (PHB) Content in Ralstonia eutropha Using Gas Chromatography and Nile Red Staining. Bio-protocol 8(5): e2748. DOI: 10.21769/BioProtoc.2748.
Category
Microbiology > Microbial biochemistry > Other compound
Biochemistry > Other compound > Poly-β-hydroxybutyrate
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









