- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fish Bile Clean-up for Subsequent Zymography and Mass Spectrometry Proteomic Analyses
Published: Vol 8, Iss 2, Jan 20, 2018 DOI: 10.21769/BioProtoc.2706 Views: 7978
Reviewed by: Samik BhattacharyaPrashanth N SuravajhalaSalome Calado Botelho

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Immunocomplexes from Zebrafish Brain
Jennifer Carlisle Michel and Adam C. Miller
Apr 5, 2023 1377 Views

A One-Step Method for Efficient Purification of Functional Cas9 Protein
Xinzhi Duan [...] Aihua Mao
Feb 5, 2026 63 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 38 Views
Abstract
Biliary excretion offers a way to analyze various contaminants in aquatic organisms, and fish bile has been used as a biomarker for environmental contamination. The use of the fish bile proteome as a tool for monitoring the impact of environmental contaminants has been recently validated. However, scarce studies in this context are available, and much remains to be investigated. In this context, this protocol describes a fast, reproducible and cheap biliary clean-up procedure for subsequent proteomic analyses, such as zymography and mass spectrometry.
Keywords: Fish bileBackground
Biliary excretion is an alternative way to analyze several chemical pollutants in aquatic organisms. Fish bile has been routinely applied as a biomarker for environmental contamination for several decades, although the focus until recently has been only the detection of environmental contaminants, and not the evaluation of possible biochemical effects, such as differential protein expression. However, the fish bile proteome has been recently validated as a tool for monitoring the impact of environmental contaminants on fish metabolism. These include the possible deleterious effects of polycyclic aromatic hydrocarbons (Pampanin et al., 2014), metals and metalloids (Hauser-Davis et al., 2012a) and complex mixtures (Hauser-Davis et al., 2012b), among other pollutants. However, this is a very recent field, and studies are still lacking in this regard. In this context, fish bile contains high amounts of lipids and bile salts, which interfere in protein quantification, 1D- and 2D-electrophoresis and mass spectrometry analyses (Hauser-Davis et al., 2012b). Thus, clean-up procedures are of the utmost importance in order to obtain reproducible results and adequately prepare this fluid for subsequent proteomic applications.
Materials and Reagents
- Pipette tips (Corning, Axygen®, catalog number: T-1005-WB-C )
- 2 ml polypropylene microcentrifuge tubes (Corning, Axygen®, catalog number: MCT-200-C )
- Sterile syringes (3 ml or higher)
- Vivaspin 500 concentrators, MWCO 3 kDa (Sartorius, catalog number: VS0191 )
- Fish
- Alcohol (100%)
- Cleanascite® (Biotech Support Group, catalog number: X2555-50 )
Equipment
- Stainless steel scissors
- Pipette 100-1,000 µl (Eppendorf, catalog number: 3120000267 )
- Ultrasonic water bath (DK SONIC, model: DK-80 )
- Refrigerated centrifuge (Eppendorf, model: 5415 R )
- Orbital shaker (Wincom/OEM/Neutral, model: TS-2000A )
Procedure
A flowchart describing the biliary clean-up procedure is displayed below in Figure 1.
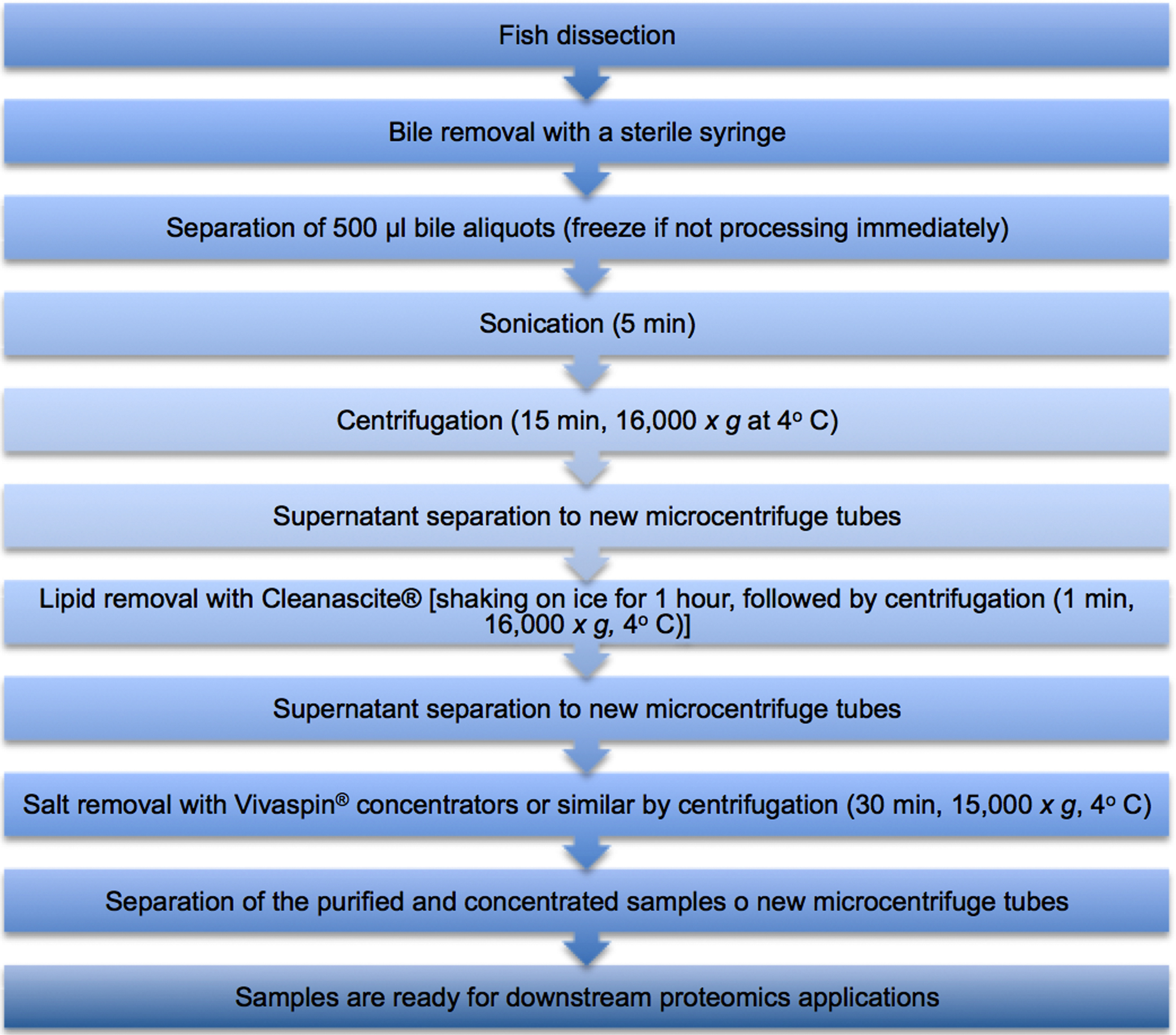
Figure 1. Flowchart describing the biliary clean-up procedure
- Fish dissection procedure
- Before each dissection, the stainless steel scissors should be cleaned with alcohol (100%) to avoid contamination.
- An incision is made from the urogenital pore up to the gills using a stainless steel scissors. After the incision is made, the operator, using both hands, may split open the fish to expose the internal organs, more specifically, the liver.
- After locating the liver, the operator should move his/her fingers behind this organ, in order to locate the gallbladder, without causing any mechanical damage to this organ (Figure 2). It is important to note that some fish species either do not possess a gallbladder, or it is too small to collect an adequate bile volume.
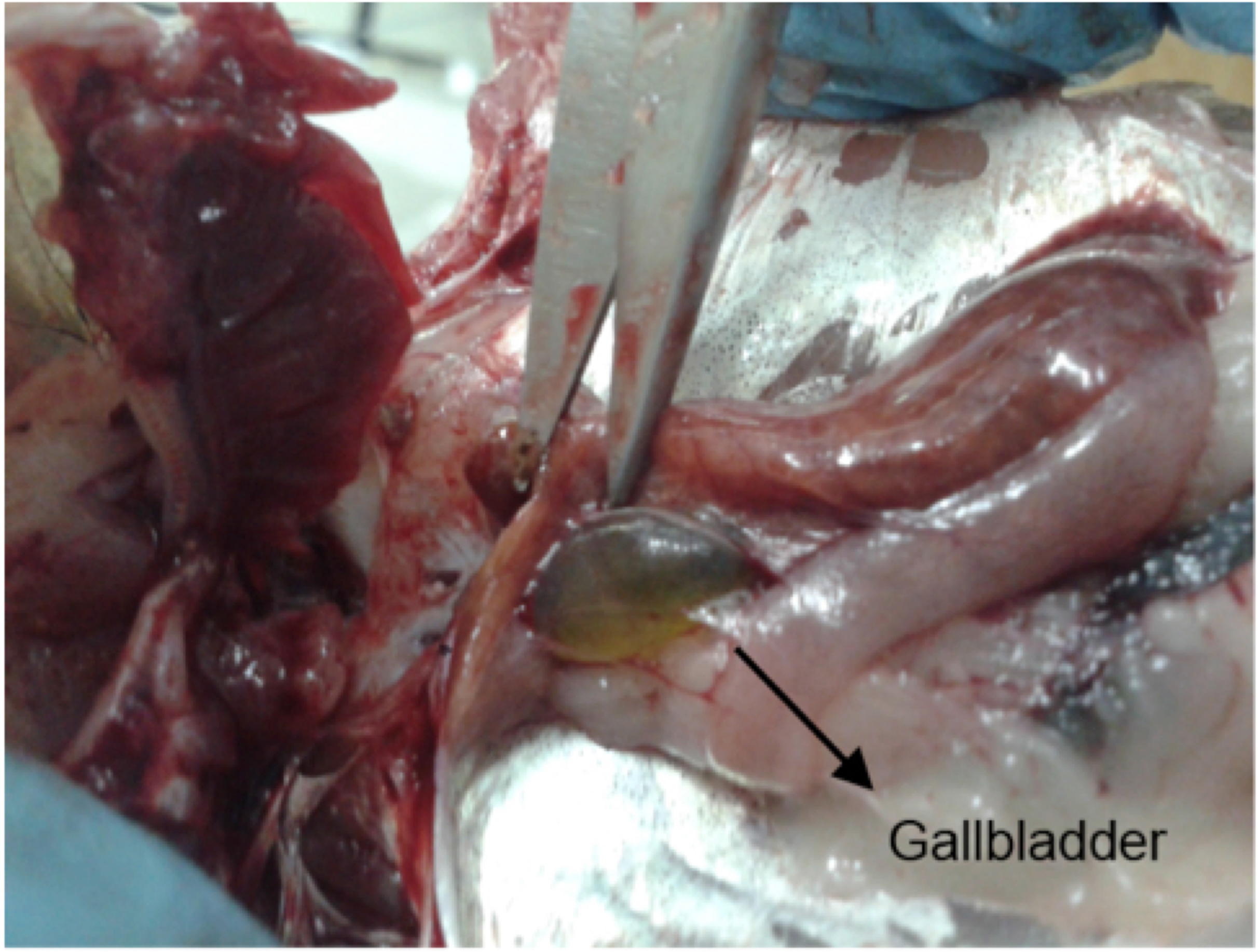
Figure 2. Fish gallbladder, pointed out by the scissors tip, usually located behind the liver. The gallbladder in question is full, with light green bile.
- After locating the gallbladder, sterile syringes are used to puncture the gallbladder and obtain the biliary fluid. Remove as much of the fluid as possible.
- Bile color and volume should be recorded, and the samples should immediately be aliquoted into 500 μl aliquots in sterile 2 ml polypropylene microcentrifuge tubes and frozen if not processed at once (preferably, at -80 °C. If an ultra-freezer is not available, samples may be frozen at -20 °C).
- Before each dissection, the stainless steel scissors should be cleaned with alcohol (100%) to avoid contamination.
- Bile processing
- Bile samples are sonicated for 5 min in order to break down cellular membranes. The use of a cold water-bath is preferred. If one is not available, the microcentrifuge tube should be removed every 20-30 sec from the bath, to avoid over-heating.
- After sonication, the samples are centrifuged for 15 min at 16,000 x g (4 °C), in order to remove cellular debris. There is no need to pre-cool the microcentrifuge tubes, since centrifugation is conducted at 4 °C and the next step is conducted on ice.
- After the centrifugation, transfer the sample supernatants to new sterile 2 ml polypropylene microcentrifuge tubes. Probably no pellet will be visible, only a slight slick film stuck to the tube wall (Step B2). Separate the supernatant anyway.
- The samples are then delipidized to avoid interferences in downstream proteomic applications.
The commercial solid-phase, non-ionic adsorbent lipid removal agent, Cleanscite® is used for this purpose, according to manufacturer’s instructions. This consists of adding a total of 75 μl of this reagent to 300 μl of the sample. (These volumes may be reduced, but the Cleanscite® solution:sample ratio should be maintained). It is important to shake the Cleanscite® solution prior to application, since the detergent may have slightly solidified.
- After adding the Cleanscite®, samples are shaken on an orbital shaker, on ice, for 1 h and then centrifuged for 1 min at 16,000 x g (4 °C) to precipitate the lipids.
- After centrifugation, transfer the sample supernatants to new sterile polypropylene microcentrifuge tubes. The pellet will, again, probably only be a slight slick film stuck to the tube wall (Step B5). Separate the supernatant anyway.
- The samples are then desalted, since salts interfere with downstream proteomic applications, and concentrated, by using Vivaspin® (or similar) concentrators. The molecular weight cut-off used depends on the aim of the study, in this case, 3 kDa. When using Vivaspin 500®, add a maximum volume of 500 μl of the sample to the concentrator, and centrifuge at 15,000 x g for 30 min (4 °C). Subsequently, transfer the liquid remaining at the top of the concentrator to new sterile polypropylene microcentrifuge tubes.
- Samples are ready for downstream proteomics applications, such as zymography and mass spectrometry.
- Bile samples are sonicated for 5 min in order to break down cellular membranes. The use of a cold water-bath is preferred. If one is not available, the microcentrifuge tube should be removed every 20-30 sec from the bath, to avoid over-heating.
Data analysis
The adequacy of the sample clean-up procedure can be verified by the following:
- Determine the total protein content by Bradford, Lowry or any other usual method. Before sample clean-up, inconsistent data is obtained for total protein content in bile, due to the presence of high amounts of lipids and bile salts, with extremely high replicate variability. After clean-up, replicates should be reproducible.
- Run a 1D protein SDS-PAGE electrophoresis or zymogram gel to check for smearing (see Hauser-Davis et al. [2012b], Figure 3, for an example visualized on a 10 % gelatin zymogram of an inadequate sample, displaying extreme smearing and few apparent lytic bands, and Figures 4 and 5 for adequate results).
Notes
- Fish should be obtained as fresh as possible, to avoid proteolytic degradation of the bile samples.
- Bile volume and color should be recorded if inferences regarding food status and exposure time are desired (Hauser-Davis, 2017).
- A avoid repeated freeze-thaw cycles of the samples (not more than 5 times), as usual in proteomic applications, in order to reduce irreproducibility.
Acknowledgments
This protocol is adapted from Hauser-Davis et al. (2012b). There are no conflicts of interest or competing interests. The author would like to thank the Brazilian National Council for Scientific and Technological Development (CNPq) for financial support.
References
- Hauser-Davis, R. A. (2017). Characterization of biliary caseinolytic proteases in two environmental contamination sentinel fish species with an emphasis on metalloproteinases. In: Daniels, J. A. (Ed.). Advances in Environmental Research. Volume 55 pp: 101-126.
- Hauser-Davis, R. A., Goncalves, R. A., Ziolli, R. L. and de Campos, R. C. (2012a). A novel report of metallothioneins in fish bile: SDS-PAGE analysis, spectrophotometry quantification and metal speciation characterization by liquid chromatography coupled to ICP-MS. Aquat Toxicol 116-117: 54-60.
- Hauser-Davis, R. A., Lima, A. A., Ziolli, R. L. and Campos, R. C. (2012b). First-time report of metalloproteinases in fish bile and their potential as bioindicators regarding environmental contamination. Aquat Toxicol 110-111: 99-106.
- Pampanin D. M., Larssen E., Øysæd K. B., Sundt R. C. and Sydnes, M. O. (2014). Study of the bile proteome of Atlantic cod (Gadus morhua): Multi-biological markers of exposure to polycyclic aromatic hydrocarbons. Mar Environ Res 101: 161-168.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hauser-Davis, R. A. (2018). Fish Bile Clean-up for Subsequent Zymography and Mass Spectrometry Proteomic Analyses. Bio-protocol 8(2): e2706. DOI: 10.21769/BioProtoc.2706.
Category
Biochemistry > Protein > Isolation and purification
Molecular Biology > Protein > Isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










