- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Identification and Quantification of Secondary Metabolites by LC-MS from Plant-associated Pseudomonas aurantiaca and Pseudomonas chlororaphis
Published: Vol 8, Iss 2, Jan 20, 2018 DOI: 10.21769/BioProtoc.2702 Views: 12260
Reviewed by: Neelanjan BoseGiovanna SalbitaniJohn F. C. Steele

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

β-lactamase (Bla) Reporter-based System to Study Flagellar Type 3 Secretion in Salmonella
Fabienne F. V. Chevance and Kelly T. Hughes
Jun 20, 2023 1764 Views

Determination of Poly(3-hydroxybutyrate) Content in Cyanobacterium Synechocystis sp. PCC 6803 Using Acid Hydrolysis Followed by High-performance Liquid Chromatography
Janine Kaewbai-ngam [...] Tanakarn Monshupanee
Aug 20, 2023 1808 Views

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1779 Views
Abstract
Increased antibiotic resistance of plants and human pathogens and continuous use of chemical fertilizers has pushed microbiologists to explore new microbial sources as potential antagonists. In this study, eight strains of Pseudomonas aurantiaca and Pseudomonas chlororaphis, have been isolated from different plant sources and screened for their antagonistic and plant growth promoting potential (Shahid et al., 2017). All strains were compared with reference strain PB-St2 and their secondary metabolites were isolated by the use of solvent partitioning and subjected to LC/ESI/MS for confirmation of compounds. The ESI-mass spectra obtained were used to characterize the surfactants ionization behavior and [M + H]+ and [M + Na]+ ions were monitored for phenazines, derivatives of lahorenoic acid and cyclic lipopeptide (WLIP). LC-MS and HPLC methods were developed to see the elution of dominant metabolites in a single run to avoid the labor and separate methods of detection for all compounds. The method was found suitable and distinctively separated the compounds at different retention times in gradient flow. This method can be helpful to explore the metabolome of Pseudomonas sp. overall and in identification and quantification of strain specific metabolites.
Keywords: PhenazinesBackground
Authors in this research group are working on bioformulations and have isolated a large number of bacteria, including pseudomonads with potential plant growth promoting abilities. Pseudomonads are known for their biocontrol ability through antibacterial and antifungal secondary metabolites. These secondary metabolites successfully counter several phytopathogens of economically important crops and Pseudomonas based biofertilizers developed so far, have been marketed as commercial products (Mandryk et al., 2007; Rovera et al., 2014). Pseudomonas aurantiaca and P. chlororaphis strains are well known biocontrol agents due to the production of phenazines and cyclic lipopeptides and their broad spectrum antagonistic activities. Although many compounds have been reported by Pseudomonas aurantiaca and P. chlororaphis to date, the list of unidentified compounds is long (Chin et al., 2001; Hu et al., 2014).
Major metabolites produced by our strains were, white line inducing principle (WLIP), 2-hydroxy phenazine (2-OH-PHZ), phenazine-1 carboxylic acid (PCA) and Lahorenoic acid A. Previously reported methods for the detection of these compounds usually account only for HPLC, and comparison of results with standards. Moreover, detailed information about their MS/MS spectra and LC-MS methods is not available. Many researchers described the short HPLC runs for the detection of phenazines that did not account for the metabolites that elute at later stages (El-sayed et al., 2008; Upadhyay and Srivastava, 2011).
Lahorenoic acid A was exclusively reported by our group (Mehnaz et al., 2013) and since then has not been reported by any other group. WLIP was also reported first time from Pseudomonas aurantiaca PB-St2 in the same paper, though it has been reported earlier from other species of Pseudomonas (Meng et al., 2014). Keeping these findings in mind, a method was established that accounts for all the basic details in detection of these compounds from Pseudomonas aurantiaca in particular and other pseudomonads in general. LC-MS method was established with long run time and gradient flow, to make the better separation of these compounds. For confirmation of results, MS/MS fragmentation patterns of all compounds were also observed. The same LC-MS method was used for HPLC detection of these compounds, however, a different column was used. Many research groups working on metabolomics do not have access to standards of every compound. It was difficult to get the standards by our group as well and therefore, purified fractions of these compounds manually collected through HPLC, were used as reference standards for relative quantification of detected compounds. This method is significant as it provides the details about four essential and main secondary metabolites of Pseudomonas sp. in a single LC-MS and HPLC run.
Materials and Reagents
- Pipette tips (AxygenTM 1,000 μl Universal Pipette Tips) (Corning, Axygen®, catalog number: T-1000-B )
- Sterile syringe filters of 0.2 µm (Corning, catalog number: 431225 )
- Pasteur pipettes (e.g., FisherbrandTM disposable borosilicate glass Pasteur pipettes, Fisher Scientific, catalog number: 13-678-20A )
- pH indicator strips (pH 0-14 Universal Indicator) (Merck, catalog number: 1095350001 )
- Nine bacterial strains including eight isolates of Pseudomonas chlororaphis subsp. aurantiaca (GS-1, GS-3, GS-4, GS-6, GS-7, ARS-38, PBSt-2, FS-2) and one Pseudomonas chlororaphis subsp. chlororaphis (RP-4) isolate
- Hydrochloric acid (HCl), 36.5-38% (Sigma-Aldrich, catalog number: 320331 )
- Ethyl acetate, EMSURE® ACS, ISO, Reag. Ph Eur. (Merck, catalog number: 1096232500 )
- Sodium sulfate, anhydrous, ≥ 99% (Sigma-Aldrich, catalog number: 1614807 )
- Methanol gradient grade for liquid chromatography, LiChrosolv® Reag. Ph Eur (Merck, catalog number: 1060072500 )
- Chloroform, EMSURE® ACS, ISO, Reag. Ph Eur. (Merck, catalog number: 1024452500 )
- Water, LC-MS grade, LiChrosolv® (Merck, catalog number: 1153332500 )
- Dimethyl sulfoxide (DMSO), ≥ 99% (Sigma-Aldrich, catalog number: 472301 )
- Formic acid, 98-100%, EMSURE® ACS, ISO, Reag. Ph Eur. (Merck, catalog number: 1002640510 )
- Acetonitrile gradient grade for liquid chromatography, LiChrosolv® Reag. Ph Eur (Merck, catalog number: 1000302500 )
- Protease-peptone, microbiology grade (Sigma-Aldrich, catalog number: 82450 )
- Glycerol, molecular biology grade, ≥ 99.5% (Sigma-Aldrich, catalog number: G9012 )
- Potassium phosphate dibasic (K2HPO4), ACS reagent, ≥ 98% (Sigma-Aldrich, catalog number: P3786 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O), ACS reagent, ≥ 98% (Sigma-Aldrich, catalog number: 230391 )
- Agar (for solid medium only), powder for Microbiology (Sigma-Aldrich, catalog number: 05040 )
Equipment
- Microbiological static incubator (e.g., Memmert, model: IF55 standard incubator )
- Microbiological incubator shaker (e.g., IKA, model: KS 4000 i control )
- Pipettes (e.g., Sartorius, catalog numbers: 728020 , 728050 , 728060 and 728070 )
- Large-volume centrifuge (e.g., HERMLE Labortechnik, model: Centrifuge ZK 496 )
- Erlenmeyer flasks (e.g., PYREXTM, 1,000 ml, 500 ml, Corning, PYREX®, catalog number: 4980-1L )
- Separatory funnels with stopcock and ring stand (e.g., 1,000 ml, 500 ml, VWR, catalog number: 89426-640 )
- Autoclave
- Glass beakers (e.g., PYREXTM, 1,000 ml, 500 ml, 100 ml, 50 ml, DWK Life Sciences, KIMBLE, catalog number: 14000-2000 )
- Glass stirrer
- Rotary evaporator (e.g., Buchi Rotavapor® R-100 (BÜCHI Labortechnik, model: Rotavapor® R-100 ) with Re-circulating Chiller F-100/F-105 (BÜCHI Labortechnik, model: Recirculating Chiller F-100/F-105 ), Vacuum Pump V-100 (BÜCHI Labortechnik, model: Vacuum Pump V-100 ), water bath and Interface I-100 (BÜCHI Labortechnik, model: Interface I-100 ))
- Fume hood (e.g., Labconco, model: 4’ Protector XStream Laboratory Hood, catalog number: 110410000 )
- LC-MS (e.g., Thermo Finnegan LC-MS, ESI Ion Trap Mass Spectrometer, Thermo Electron, model: LCQ Advantage Max )
- TLC plates (Silica Gel 60G F254 20 x 20 cm, e.g., Merck, catalog number: 100390 )
- C18 column for MS (e.g., Thermo Hypersil Gold C18 column, Length 250 mm, 4.6 mm ID, 5 µm particle size, Thermo Fisher Scientific, catalog number: 25005-254630 , Lot No. 7648, S/N 0871381M)
- HPLC system (e.g., WATERS, model: e2995 Separations Module , 2998 Photodiode Array (PDA) Detector)
- HPLC column (e.g., Nucleosil C18 column, 4.6 x 250 mm, 5 µM; Macherey-Nagel, Germany)
Software
- Xcalibur 2.0 software (XcaliburTM Software, control and process data from LC-MS instruments. It is WindowsTM based software that provides method setup, data acquisition, data processing and reporting. Data files are retrieved in Qual Browser and are processed for interpretation and analysis)
Procedure
- Culture conditions
- Total nine bacterial strains including eight isolates of Pseudomonas chlororaphis subsp. aurantiaca (GS-1, GS-3, GS-4, GS6, GS-7, ARS-38, PBSt-2, FS-2) and one isolate Pseudomonas chlororaphis subsp. chlororaphis (RP-4) were included in this research. These bacterial strains are 16S rRNA identified and screened for their PGPR activities (Shahid et al., 2017). All bacterial strains were streaked on King’s B agar plates (see Recipes) and plates were incubated at 28 ± 2 °C for 48 h.
- A single colony of each bacterial strain; GS-1, GS-3, GS-4, GS6, GS-7, ARS-38, PBSt-2, FS-2 and, RP-4 was separately used to inoculate 10 ml of King’s B broth. Cultures were incubated in a shaking incubator for 24 h at 150 rpm and 28 ± 2 °C. Next day, each bacterial culture was individually inoculated (2%) in 500 ml King’s B broth and flasks were incubated at 28 ± 2 °C, 150 rpm for 96 h.
- Total nine bacterial strains including eight isolates of Pseudomonas chlororaphis subsp. aurantiaca (GS-1, GS-3, GS-4, GS6, GS-7, ARS-38, PBSt-2, FS-2) and one isolate Pseudomonas chlororaphis subsp. chlororaphis (RP-4) were included in this research. These bacterial strains are 16S rRNA identified and screened for their PGPR activities (Shahid et al., 2017). All bacterial strains were streaked on King’s B agar plates (see Recipes) and plates were incubated at 28 ± 2 °C for 48 h.
- Extraction procedure
- After 96 h of incubation, harvest cultures and centrifuge at 3,376 x g for 40 min at 4 °C. Collect supernatants in separate flasks.
- All supernatants are acidified to pH 2 with 1 N HCl and pH is monitored with pH-indicator strips.
- Acidified supernatants are extracted twice with equal volume of ethyl acetate. For this, in each flask with 500 ml of acidified supernatant, add 500 ml of ethyl acetate. Put flasks in a shaking incubator for 10-15 min and then transfer materials into separatory funnels. The materials will separate into two phases, organic phase and water phase. The upper phase is organic phase while the lower phase has culture supernatant. Collect them into separate beakers. Re-extract the collected culture supernatant with ethyl acetate and combine the re-extracted organic layer into the previous collected organic phase.
- Add 20-30 g of anhydrous sodium sulphate to the beaker with organic phase and stir with a glass stirrer. This step is essential to remove any moisture from the organic layers. Then, transfer this content to another clean and dry beaker (avoiding any salt particles) and finally in the rotary evaporator flask.
- Turn on all basic units of rotary evaporator including central interface, glass assemblies, water bath, chiller, and vacuum. Set water bath temperature to 40-45 °C, and adjust rotary rotations accordingly to prevent any bumping of liquid in glass assemblies. When the liquid phase is completely dried, separate the round bottom flask with dried residue from rotary evaporator glass assembly.
- Re-dissolve residues of extracts in methanol and chloroform (2 ml methanol:2 ml chloroform), completely dry in a fume hood and store at 4 °C.
- After 96 h of incubation, harvest cultures and centrifuge at 3,376 x g for 40 min at 4 °C. Collect supernatants in separate flasks.
- Identification of bacterial compounds using LC-MS
- For characterization of bacterial secondary metabolites, dissolve the extracts in 1.5 ml of methanol and 500 µl of chloroform.
Note: Please be sure that extracts are completely dissolved and no un-dissolved residue left in the vials. If any portion of extracts remains undissolved, collect the methanol and chloroform soluble part in separate vials and dissolve the remaining undissolved part in DMSO (Dimethyl sulfoxide). - Take 500 µl of these extracts and individually filter with sterile syringe filters of 0.2 µm (Fisher Scientific).
- Subject the extracts of all strains to Liquid Chromatography-Electrospray Ionization-Mass Spectrometry (LC/ESI/MS), for identification of secondary metabolites.
- Set up the instrument and perform LC-ESI MS/MS runs using a Thermo Finnegan HPLC system, coupled to an LCQ Advantage Max ESI-Ion Trap Mass Spectrometer (Thermo Electron, USA).
- Chromatographic separations are achieved using Thermo Hypersil Gold C18 column (4.6 x 250 mm, 5 µm particle size). Set the temperature of the column compartment at 25 °C, and load 20 µl of injection volume on the column.
- A gradient used to separate the metabolites consists of two solvent systems. Solvent A is 0.1% formic acid in water and solvent B is 0.1% formic acid in acetonitrile. Total LC-MS/MS runtime is 55 min, and set the flow rate at 0.7 ml/min.
- For characterization of bacterial secondary metabolites, dissolve the extracts in 1.5 ml of methanol and 500 µl of chloroform.
- Gradient conditions of LC-MS/MS
- Set gradient conditions as follows (Table 1):
Table 1. Gradient run used for LC-MS and HPLC analyses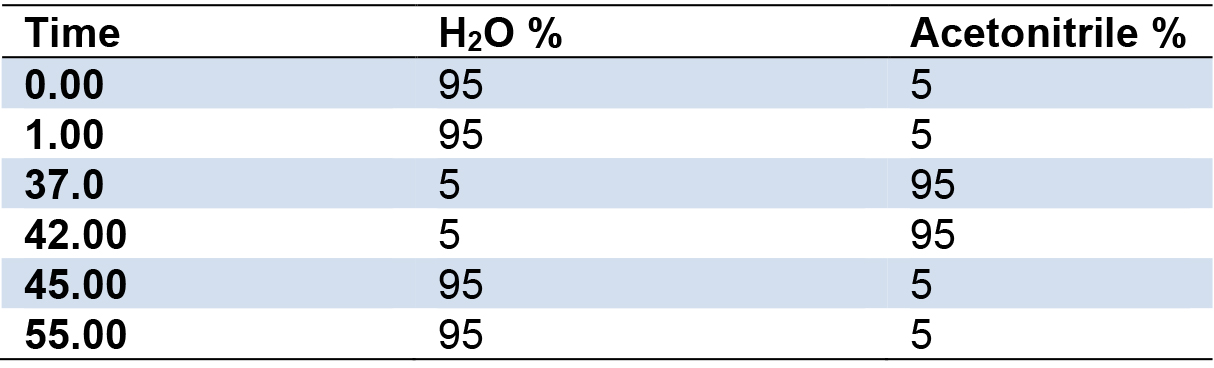
- ESI positive mode is used for the runs with data dependent protocol with a mass range of 150-1,500 a.m.u. and two scan events are employed for this data. The first scan event is a full scan of 150-1,500 and the second scan is dependent on the most abundant ions in the first scan triggering their MS2 acquisition.
- Data are acquired at the normalized collision energy of 30%. The heated capillary is maintained at 350 °C, and sheath and auxiliary/sweep gases are at 60 and 25 arbitrary units, respectively.
- Set the source voltage to 4.5 kV with 10 V capillary voltage. The ESI-mass spectra obtained are used to characterize the surfactant ionization behavior; [M + H]+ and [M + Na]+ ions are monitored for phenazines, i.e., 2-hydroxy-phenazine(2-Oh-Phz) and phenazine-1-carboxylic acid (PCA); Lahorenoic acid A, and cyclic lipopeptide (WLIP). In addition, the ESI-MS/MS fragmentation behavior of identified peaks is investigated to confirm the structure of these secondary metabolites. Figures (Figures 1-3 and Supplemental Figures S1-S10) describe the structures, extracted ion current (XIC) chromatograms, mass spectrums and MS/MS fragmentation behavior of detected metabolites. Chemical formulas, exact masses and observed m/z values for detected metabolites have been given in Table 2.
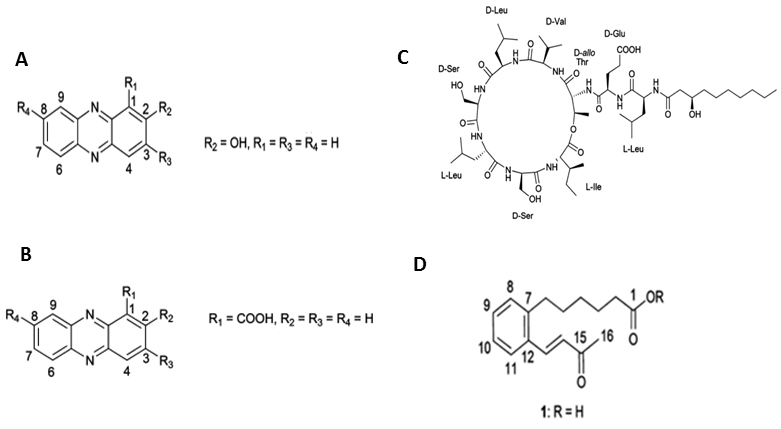
Figure 1. Structures of the compounds isolated from Pseudomonas aurantiaca and Pseudomonas chlororaphis. A. 2-hydroxyphenazine, B. phenazine-1-carboxylic acid, C. white-line-inducing principle, D. lahorenoic acid A (Mehanz et al., 2013).
Table 2. Chemical formulas, exact masses and observed m/z values for detected metabolites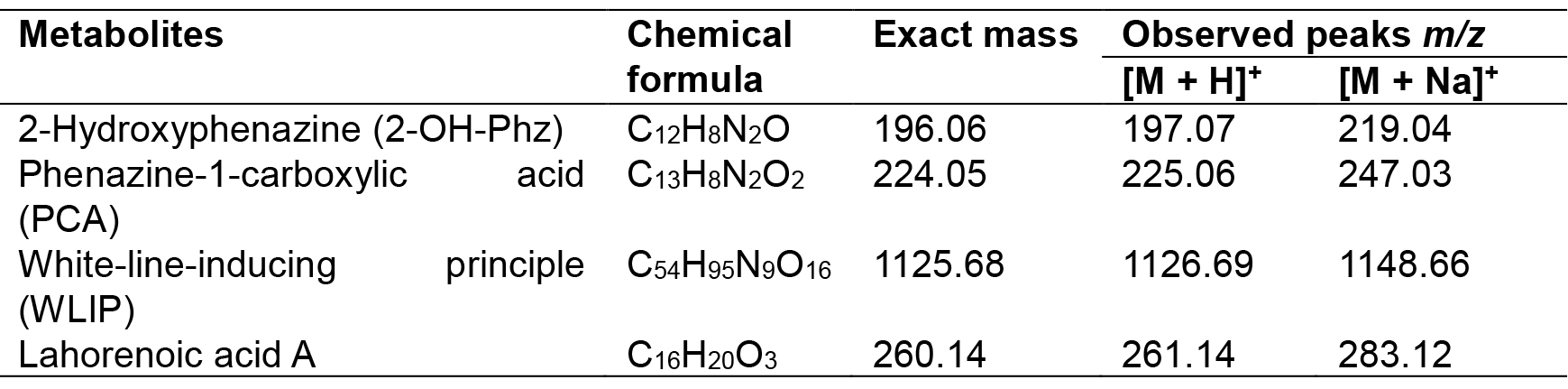
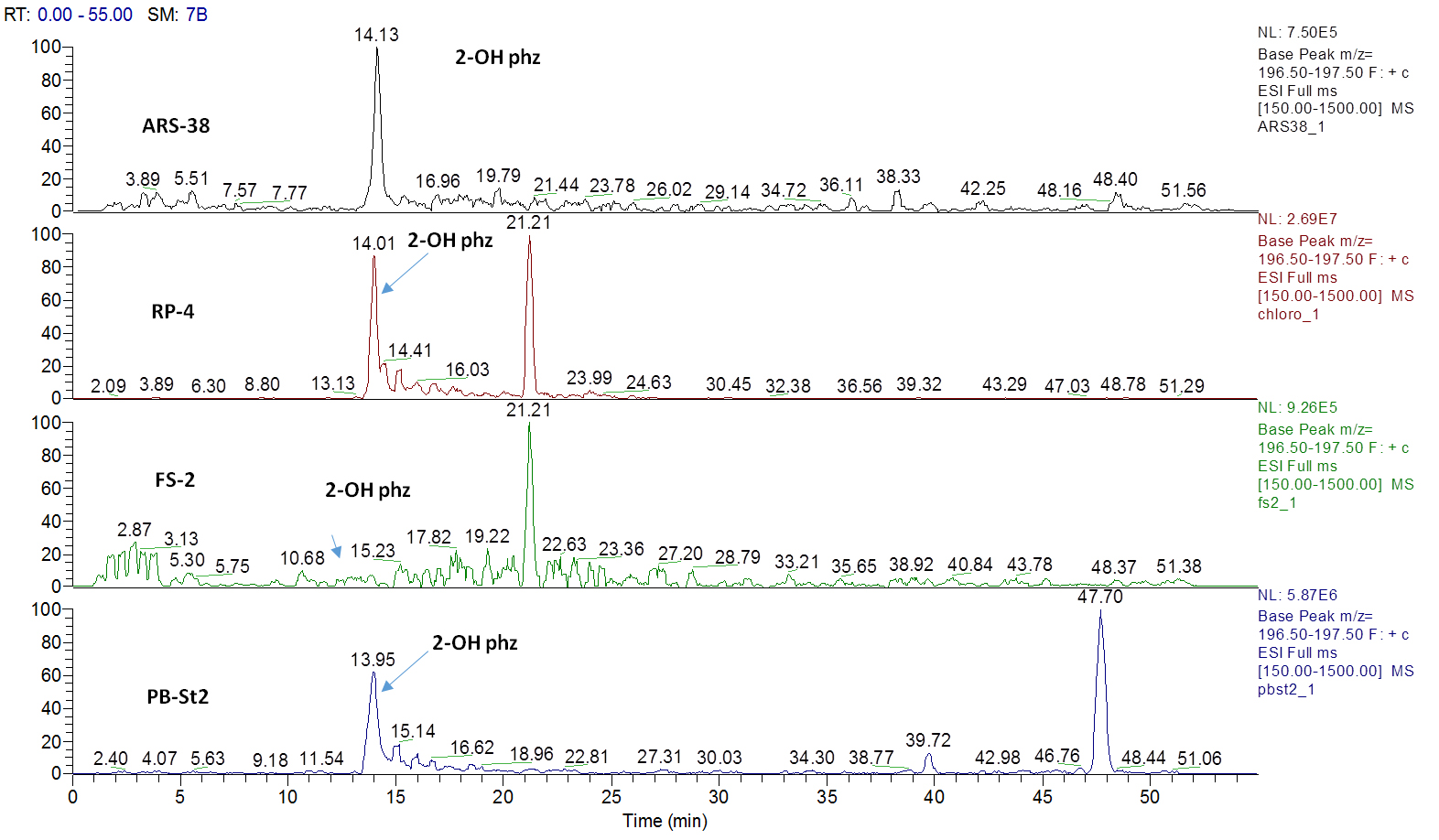
Figure 2. Extracted ion current (XIC) chromatograms for 2-hydroxyphenazine (2-OH-Phz), m/z 197, [M + H]+ of four strains; ARS-38, RP-4, FS-2 and PB-St2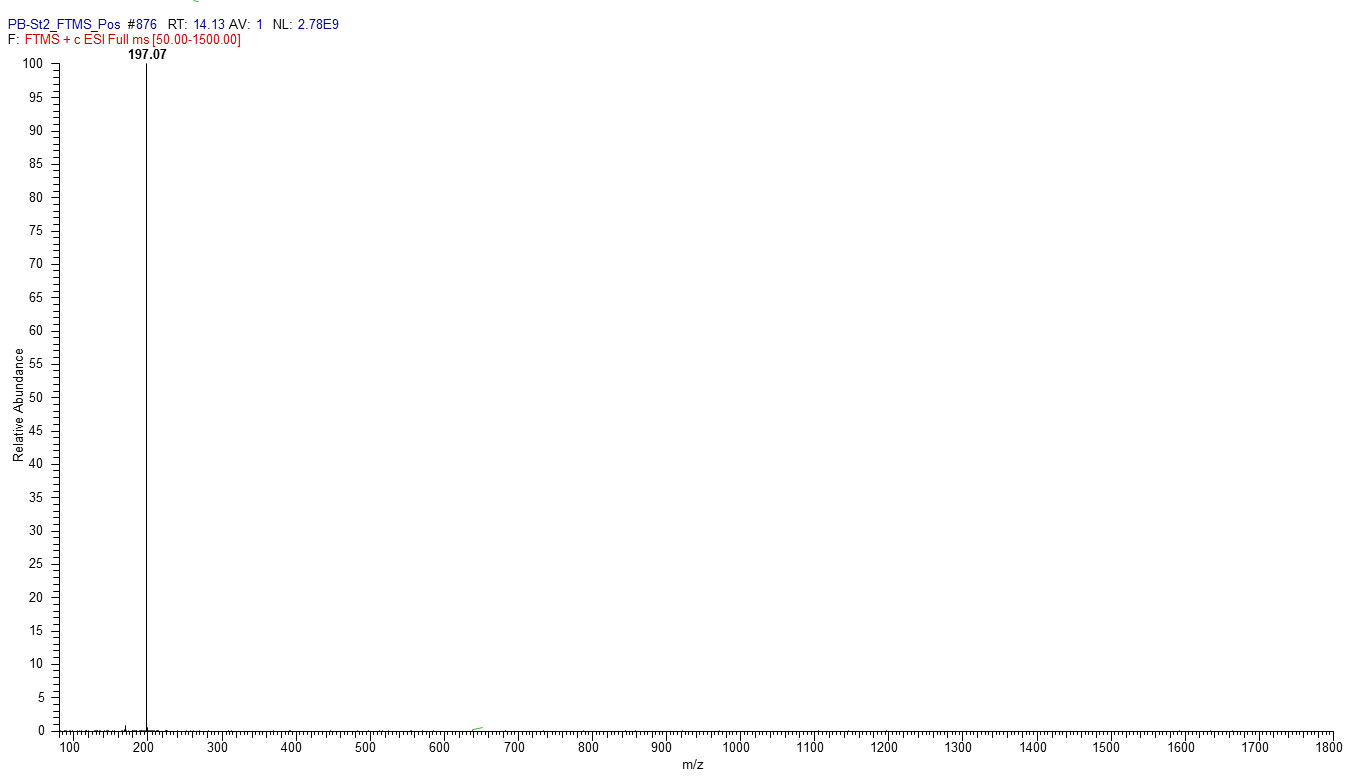
Figure 3. Mass spectrum of 2-Hydroxyphenazine, m/z [M + H] + 197.07
- Set gradient conditions as follows (Table 1):
- Thin layer chromatography (TLC) of identified secondary metabolites
Identified four secondary metabolites can also be analyzed by thin layer chromatography (TLC), using PB-St2 as a reference strain. PB-St2 is used as reference strain as all of its secondary metabolites have previously been published (Mehnaz et al., 2009 and 2013).- For thin layer chromatography, load methanol fractions of all bacterial extracts (10 µl) on TLC plates (Silica Gel 60G F254 20 x 20 cm). Mobile phase contains chloroform: acetone: acetic acid (78.4:20:1.6, v/v/v) and samples are spot inoculated on TLC plate for separation (Figure 4).
- Air dry TLC plates (approximately for 5-10 min) and analyze for showing the separation of phenazines, i.e., phenazine-1-carboxylic acid (PCA) and 2-hydroxy phenazine (2-OH-Phz).
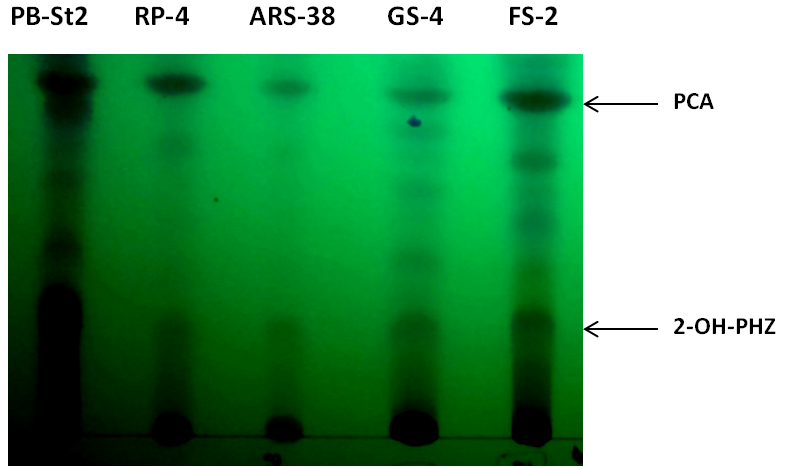
Figure 4. Thin layer chromatogram of phenazine-1-carboxylic acid (PCA) and 2-hydroxy phenazine (2-OH-PHZ), present in cell-free supernatants of bacterial extracts of RP-4, ARS-38, GS-4 and FS-2. PB-St2 fractions were used as reference standard for this TLC analysis.
- For thin layer chromatography, load methanol fractions of all bacterial extracts (10 µl) on TLC plates (Silica Gel 60G F254 20 x 20 cm). Mobile phase contains chloroform: acetone: acetic acid (78.4:20:1.6, v/v/v) and samples are spot inoculated on TLC plate for separation (Figure 4).
- Quantification of secondary metabolites
- Manually collect the pure fractions of these compounds using HPLC from PB-St2 Pseudomonas aurantiaca isolate (or any bacterium that needs to be analyzed for its secondary metabolites) and analyze on LC-MS for their purity (Figure 5). The method for LC-MS is the same as used in HPLC analysis. The total run time is 55 min, with 20 µl injection volume and with same acetonitrile and water gradient run, as described in Table 1.
- Analyze samples on Waters HPLC System (e2995, separations module) with 299 h photodiode-array (PDA) detector using a Nucleosil C18 column (4.6 x 250 mm, 5 µM; Macherey-Nagel, Germany). Collected fractions of these compounds are used as reference standard to quantify the production of these compounds from all bacteria used in this study.
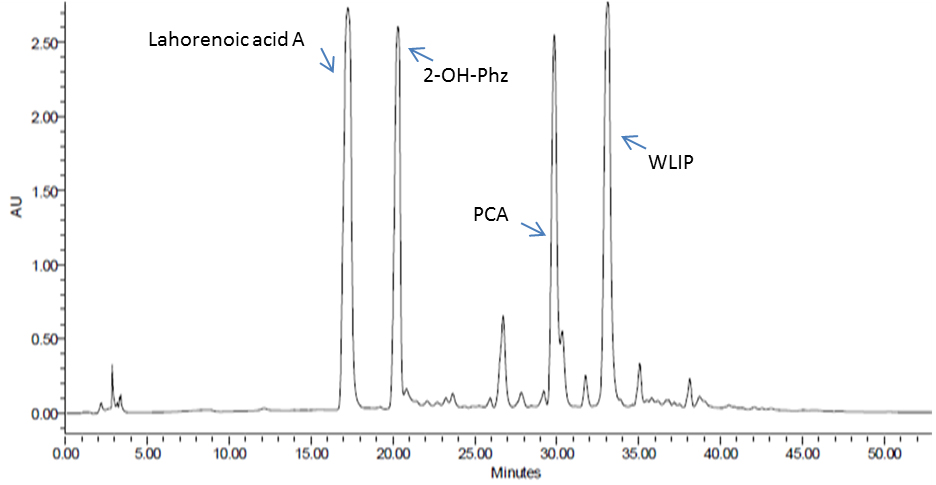
Figure 5. HPLC analysis of enriched P. aurantiaca PB-St2 fractions of detected metabolites
- Manually collect the pure fractions of these compounds using HPLC from PB-St2 Pseudomonas aurantiaca isolate (or any bacterium that needs to be analyzed for its secondary metabolites) and analyze on LC-MS for their purity (Figure 5). The method for LC-MS is the same as used in HPLC analysis. The total run time is 55 min, with 20 µl injection volume and with same acetonitrile and water gradient run, as described in Table 1.
Data analysis
All LC-MS/MS data were acquired and data files were processed using Xcalibur 2.0 software. Data files were initially open using Xcalibur FTMS raw data files. These files displayed total ion chromatogram (TIC) on the top and m/z spectra of total scan at the bottom. Using software’s basic features, data were acquired for reported four compounds and their ESI-MS/MS fragmentation patterns were also observed to confirm the structure of these secondary metabolites. ESI-MS/MS fragmentation was also confirmed with Wishart Research Group’s (CFM-ID), Competitive Fragmentation Modeling for Metabolite Identification (cfmid.wishartlab.com).
Notes
- Avoid any plastic equipment during the preparation of samples for HPLC or LC-MS and also at the time of extraction. This may add some plasticizers in your prepared samples that often result in false peaks in LC-MS and HPLC run.
- Amount of the culture medium can also be reduced accordingly. This method used the fractions extracted from 500 ml of King’s B broth. It can be reduced to 100 ml or 50 ml according to the requirement. Amount of organic solvents used will also get reduced with it.
- For extraction with ethyl acetate, the supernatant is acidified to pH 2, while using dichloromethane (DCM) for extraction of secondary metabolites, the supernatant is not acidified/pH not changed.
Recipes
- King’s B agar and broth
Protease peptone 20 g/L
Glycerol 20 g/L
Anhydrous K2HPO4 1.5 g/L
MgSO4·7H2O, 6.09 ml of 1 M solution
Agar (for solid medium only), 15 g/L
Deionized H2O, 1,000 ml
Completely dissolve protease peptone, glycerol and anhydrous K2HPO4 in 500 ml of deionized H2O and adjust the pH to 7.2. Make up the final volume to 994 ml and autoclave for 20 min. Separately, prepare 1 M solution of MgSO4·7H2O and autoclave it. Add 6.00 ml of this solution to the autoclaved medium for making up the final volume to 1,000 ml
Acknowledgments
The original article has been published in Journal of Microbiology and Biotechnology (Shahid et al., 2017). Authors gratefully acknowledge the support of Alexander Von Humboldt Foundation, Bonn, Germany, for equipment grant and Higher Education Commission (HEC; Project No. 20-3134), Pakistan, for supporting this research work. Authors declare that the research was conducted in the absence of any financial or commercial relationships that could be constructed as a potential conflict of interest.
References
- Chin,A. W. T. F., van den Broek, D., de Voer, G., van der Drift, K. M., Tuinman, S.,Thomas-Oates, J. E., Lugtenberg, B. J. and Bloemberg, G. V. (2001). Phenazine-1-carboxamideproduction in the biocontrol strain Pseudomonas chlororaphis PCL1391 is regulated by multiple factorssecreted into the growth medium. MolPlant Microbe Interact 14(8): 969-979.
- El-Sayed,W., El-Megeed, M., El-Razik, A. B., Soliman, K. H. and Ibrahim, S. A. (2008). Isolation and identification ofphenazine-1-carboxylic acid from different Pseudomonas isolates and its biological activity against Alternaria solani. Res J AgricBiol Sci. 4 (6): 892-901.
- Hu, W., Gao, Q., Hamada, M.S., Dawood, D. H., Zheng, J., Chen, Y. and Ma, Z. (2014). Potentialof Pseudomonas chlororaphis subsp. aurantiaca strain Pcho10 as a biocontrol agentagainst Fusarium graminearum. Phytopathology 104(12): 1289-1297.
- Mandryk,M. N., Kolomiets, E. I. and Dey, E. S. (2007). Characterizationof antimicrobial compounds produced by Pseudomonasaurantiaca S-1. PolJ Microbiol 56(4): 245-250.
- Mehnaz, S., Baig, D. N.,Jamil, F., Weselowski, B. and Lazarovits, G. (2009). Characterizationof a phenazine and hexanoyl homoserine lactone producing Pseudomonas aurantiaca strainPB-St2, isolated from sugarcane stem. JMicrobiol Biotechnol 19(12): 1688-1694.
- Mehnaz, S., Saleem, R. S.,Yameen, B., Pianet, I., Schnakenburg, G., Pietraszkiewicz, H., Valeriote, F.,Josten, M., Sahl, H. G., Franzblau, S. G. and Gross, H. (2013). Lahorenoicacids A-C, ortho-dialkyl-substituted aromatic acids from the biocontrol strain Pseudomonas aurantiaca PB-St2. J Nat Prod 76(2):135-141.
- Meng, J.,Fan, Y., Su, M., Chen, C., Ren, T., Wang, J. and Zhao, Q. (2014). WLIPderived from Lasiosphaera fenzlii Reich exhibits anti-tumor activity and induces cell cycle arrest through PPAR-γassociated pathways. Int Immunopharmacol 19(1): 37-44.
- Rovera, M.,Pastor, N., Niederhauser, M. and Rosas, S. B. (2014). Evaluationof Pseudomonas chlororaphis subsp. aurantiaca SR1 for growth promotion of soybean and for control of Macrophomina phaseolina. Biocont Sci Tech 24: 1012-1025.
- Shahid, I., Rizwan, M., Baig,D. N., Saleem, R. S., Malik, K. A. and Mehnaz, S. (2017). Secondarymetabolites production and plant growth promotion by Pseudomonas chlororaphis and P.aurantiaca strains isolated from cactus, cotton, and Para grass. JMicrobiol Biotechnol 27(3): 480-491.
- Upadhyay, A. and Srivastava, S. (2011). Phenazine-1-carboxylicacid is a more important contributor to biocontrol Fusarium oxysporum than pyrrolnitrin in Pseudomonas fluorescens strain Psd. Microbiol Res 166: 323-335.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shahid, I., Rizwan, M. and Mehnaz, S. (2018). Identification and Quantification of Secondary Metabolites by LC-MS from Plant-associated Pseudomonas aurantiaca and Pseudomonas chlororaphis. Bio-protocol 8(2): e2702. DOI: 10.21769/BioProtoc.2702.
Category
Microbiology > Microbial biochemistry > Other compound
Microbiology > Antimicrobial assay > Antifungal assay
Systems Biology > Metabolomics > Whole organism
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











