- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
D-serine Measurements in Brain Slices or Other Tissue Explants
Published: Vol 8, Iss 2, Jan 20, 2018 DOI: 10.21769/BioProtoc.2698 Views: 8100
Reviewed by: Pengpeng LiSalome Calado BotelhoJingli Cao

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Single-Particle Tracking of AMPA Receptor-Containing Vesicles
Victor C. Wong [...] Erin K. O’Shea
Jun 5, 2025 2260 Views

Local Iontophoretic Application for Pharmacological Induction of Long-Term Synaptic Depression
Borys Olifirov [...] Pavel Belan
Jun 5, 2025 1897 Views

Ultrafast Isolation of Synaptic Terminals From Rat Brain for Cryo-Electron Tomography Analysis
Rong Sun and Qiangjun Zhou
Sep 5, 2025 3556 Views
Abstract
D-serine is an atypical amino acid present in the mammalian body (most amino acids in the mammalian body are L-isomers) that is mostly known in neuroscience for its role as a co-agonist controlling the N-methyl D-aspartate receptor (NMDAR). D-serine levels are decreased in patients with schizophrenia and this is thought to mediate, at least in part, the hypofunction of NMDARs that is central to the glutamate hypothesis for the etiology of this neuropsychiatric disorder. D-serine detection was first established using high performance liquid chromatography, a costly and complex technique that requires high levels of expertise. But with the increasing interest in this unconventional amino acid, there is an increasing need for easier, cheaper and more accessible detection methods. Here we describe the amperometric, biosensor-based method we employed in a recent publication (Papouin et al., 2017b). It allows reliable measurement of D-serine levels from fresh tissue, such as acute brain slices, for concentrations higher than 100 nM, with minimal technical requirements.
Keywords: D-serineBackground
The N-methyl D-aspartate receptor (NMDAR) is a receptor for the neurotransmitter glutamate in the brain, spinal cord and in the peripheral nervous system such as enteric neurons. It is also found in renal tubular cells and chondrocytes. In addition to glutamate, the activation of the NMDAR requires the binding of a co-agonist on a dedicated binding site (Johnson and Ascher, 1987; Kleckner and Dingledine, 1988). The unconventional amino acid D-serine is the endogenous co-agonist of the NMDAR in numerous regions of the nervous system (see Papouin et al., 2017a). It is also found abundantly in the liver and kidneys where its degradation and excretion take place (Montesinos Guevara and Mani, 2016). D-serine is also found in the gut, where its function and origin (host metabolite or bacterial origin) are unclear. Therefore, detecting and measuring D-serine levels has become necessary in several subfields of neuroscience and other disciplines but has proven technically challenging. In the brain and spinal cord, assessing the occupancy of the NMDAR co-agonist binding site is an excellent first approach to assess D-serine levels (Papouin et al., 2012; Papouin et al., 2017b; Ferreira et al., 2017). However, major limitations of this approach are that 1) it provides little quantitative insights into the actual concentration of D-serine, 2) it is subject to a strong ceiling effect (once the co-agonist binding site is saturated, higher levels of D-serine go undetected), 3) glycine can also bind to the NMDAR co-agonist binding site and compete with D-serine, 4) this approach is subject to changes and differences in the affinity of the NMDAR co-agonist binding site, and finally 5) this method is only useful in conditions where recording NMDAR activity is technically possible. Therefore, obtaining direct measurements of D-serine has become a necessity and challenge. Two methods are currently available. The first one is electrophoresis-based, in particular high performance liquid chromatography (Papouin et al., 2017b) or capillary electrophoresis (Ferreira et al., 2017) which has been amply documented. While they provide high levels of precision and reliability, they also require expensive equipment and extensive technical expertise. The second one is based on the use of biosensors such as those developed by Sarissa (Dale et al., 2005) or by several independent labs such as Pernot et al., 2008, which requires minimal equipment or technical expertise. Biosensors usually consist of probes coated with the enzyme D-amino acid oxidase which degrades D-serine to produce electrons. They function as amperometric probes, where electrical current produced during D-serine degradation is used to measure the amount of D-serine present. We found that the use of sensors comes with pitfalls and caveats that, if not carefully avoided or controlled, can lead to aberrant measurements. Therefore, in a recent study (Papouin et al., 2017b) we developed a protocol to reliably detect D-serine levels in brain slices using D-serine biosensors from Sarissa. Here, we describe this protocol in greater detail, and in a step-by-step manner. This protocol is based on obtaining conditioned medium from brain slices and, therefore, can be easily adapted to any tissue of interest, such as the spinal cord, kidney or liver, provided that acute slices can be obtained.
Materials and Reagents
Materials
- For the ‘nest Beaker’
- Nylon tights
- Instant superglue (such as Scotch Super Glue, 3M, catalog number: AD124 )
- 15 ml tubes (such as VWR, catalog number: 89039-670 US, 525-0450 Europe)
- Disposable 6 cm diameter plastic Petri dish (such as Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 123TS1 )
- Nylon tights
- For dissection
- Large kitchen scissors or guillotine
- Straight fine scissors (such as Fine Science Tools, catalog number: 14060-11 )
- Curved spatula (such as Fine Science Tools, catalog number: 10092-12 )
- Scalpel (such as Fine Science Tools, catalog number: 91003-12 )
- Glass disposable Pasteur pipet (such as Fisher Scientific, FisherBrand, catalog number: 13-678-6A )
- Dropper bulb (such as Fisher Scientific, FisherBrand, catalog number: 03-448-25 )
- Plastic container, about 2.5 cm high and 150 ml, such as the lid of a pipet tip box or a large glass Petri dish (Cole-Parmer Instrument, catalog number: EW-34551-06 )
- Whatman paper (GE Healthcare, Whatman, catalog number: 1001-090 )
- Disposable Razor blade (such as Personna Double Edge Razor Blades [Amazon, PERSONNA, catalog number: BP9020 ])
- Large kitchen scissors or guillotine
- For conditioned medium procedure
- Disposable 6 cm diameter (or larger) plastic Petri dish (such as Thermo Fisher, catalog number: 123TS1 )
- 1.7 to 2 ml microtubes (such as Sorenson BioScience, catalog number: 16070 )
- Disposable 6 cm diameter (or larger) plastic Petri dish (such as Thermo Fisher, catalog number: 123TS1 )
- For biosensors holders
- 2 ml Falcon pipette (Fisher Scientific, catalog number: 13-678-11C )
- Heat-Shrink tubes (from RadioShack)
- 2 ml Falcon pipette (Fisher Scientific, catalog number: 13-678-11C )
Reagents
- Glucose (Sigma-Aldrich, catalog number: G7021 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Sodium phosphate monobasic anhydrous (VWR, catalog number: 470302-666 )
Manufacturer: ALDON, catalog number: SS0756-500GR . - Sodium bicarbonate (NaHCO3) (Sigma-Aldrich, catalog number: S5761 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333 )
- Magnesium chloride solution (1 M) (Sigma-Aldrich, catalog number: 63069 )
- Calcium chloride solution (1 M) (Sigma-Aldrich, catalog number: 21115 )
- D-serine (Sigma-Aldrich, catalog number: S4250 )
- Stock artificial cerebrospinal fluid (ACSF) solution (see Recipes)
- Ice-cold Slicing ACSF stock solution (see Recipes)
- Recovery ACSF stock solution (see Recipes)
- Experimental ACSF stock solution (see Recipes)
Equipment
- 250 ml Pyrex beaker (such as VWR, catalog number: 10754-952 )
- Straight spring scissors (such as Fine Science Tools, catalog number: 15018-10 )
- Curved fine forceps (such as Fine Science Tools, catalog number: 11152-10 )
- 600 ml Pyrex beaker (such as VWR, catalog number: 10754-956 )
- 95% O2/5% CO2 tank (such as AirGas, catalog number: Z02OX9522000043 )
- Vibratome (such as Leica, model: Leica VT 1200 S , catalog number: 14048142066)
- Bath heater (such as Thermo Fisher Scientific, Thermo Scientific, model: Precision 180 , catalog number: 51221073)
- Digi IVY DY2023 Bipotentiostat (Digi-Ivy, model: DY2023 ) (www.digi-ivy.com)
- Connecting cables (provided with bipotentiostat)
- Computer connected to the bipotentiostat
- Sarissa Probe D-serine biosensor (Sarissa Biomedical, catalog number: SBS-DSER-05-50 )
- Sarissa Probe Null Sensor (Sarissa Biomedical, catalog number: SBS-NUL-05-50 )
- Minimal components of a classic electrophysiology rig (Figure 1):
Perfusion/immersion chamber (such as Warner Instruments, model: RC-26G , catalog number: 64-0235)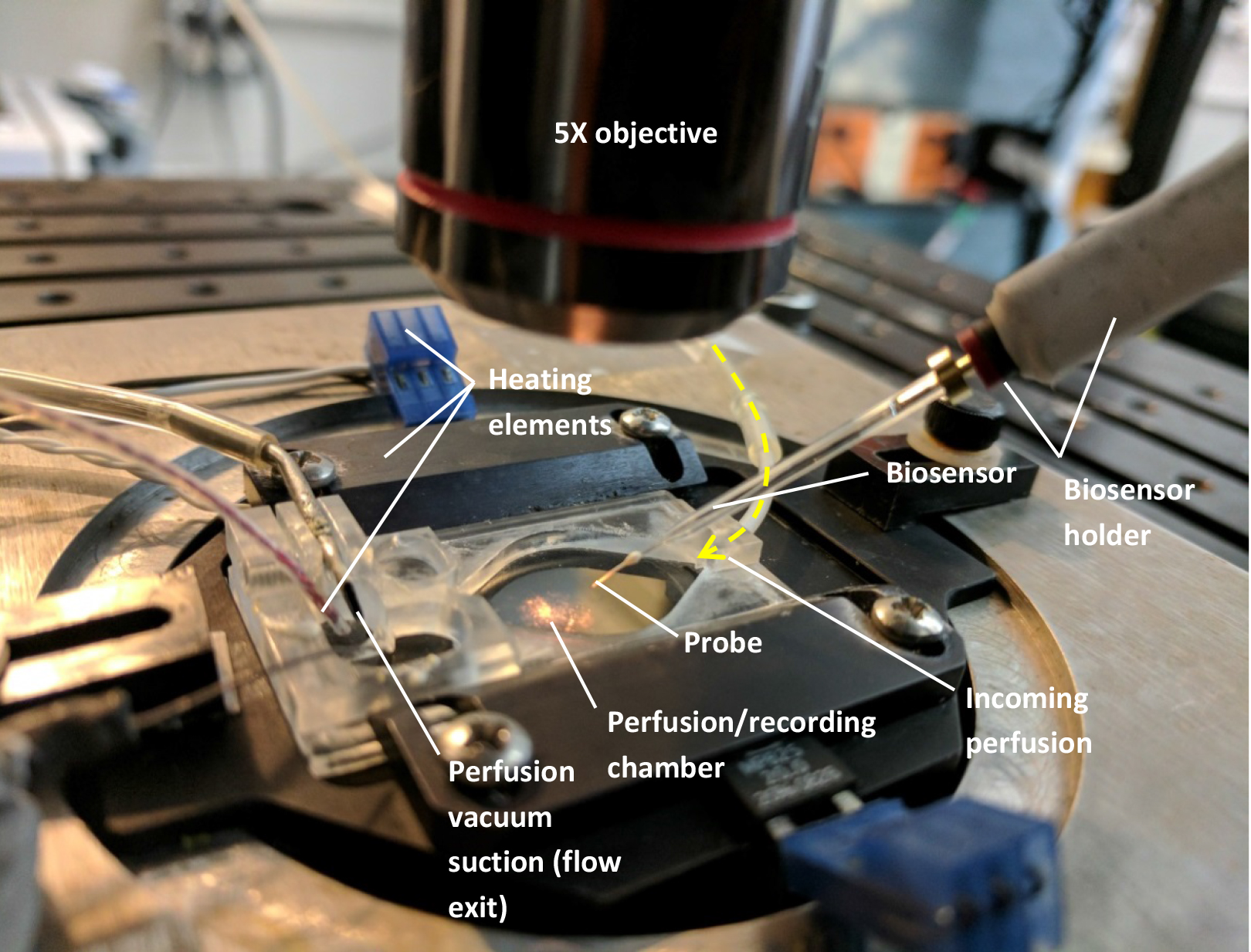
Figure 1. Setup needed to mount the biosensors (see text). For clarity, there is no liquid perfused through the chamber on the above image and only one biosensor was mounted. - Custom-made perfusion line (gravity or pump-operated) using flexible plastic tubing (such as Cole-Parmer Instrument, Masterflex, catalog numbers: ZM-96400-13 and ZM-96400-14 )
Note: If the perfusion system is gravity-based, a vacuum line connected to the exit end of the chamber will be required to suction the liquid from the chamber and maintain a steady state immersion level in the chamber.- Chamber temperature controller (such as Warner Instruments, model: TC-344C , catalog number: 64-2401)
- Microscope or any time of magnifying binocular allowing a 5x magnification (and lighting)
- Manual manipulators to allow the placement of the sensors (such as NARISHIGE, model: M-152 )
- Home-made connector to plug the sensors to the cables provided with the bipotentiostat and allowing placement in the manipulators (see Figure 2)
- Biosensor holder. Using a 2 ml Falcon pipette, cut the desired length (we recommend 10 cm), insert the biosensor connecting cable, wrap in heat-shrink tube (from RadioShack) as shown in Figure 2 and gently heat with a lighter or a soldering iron
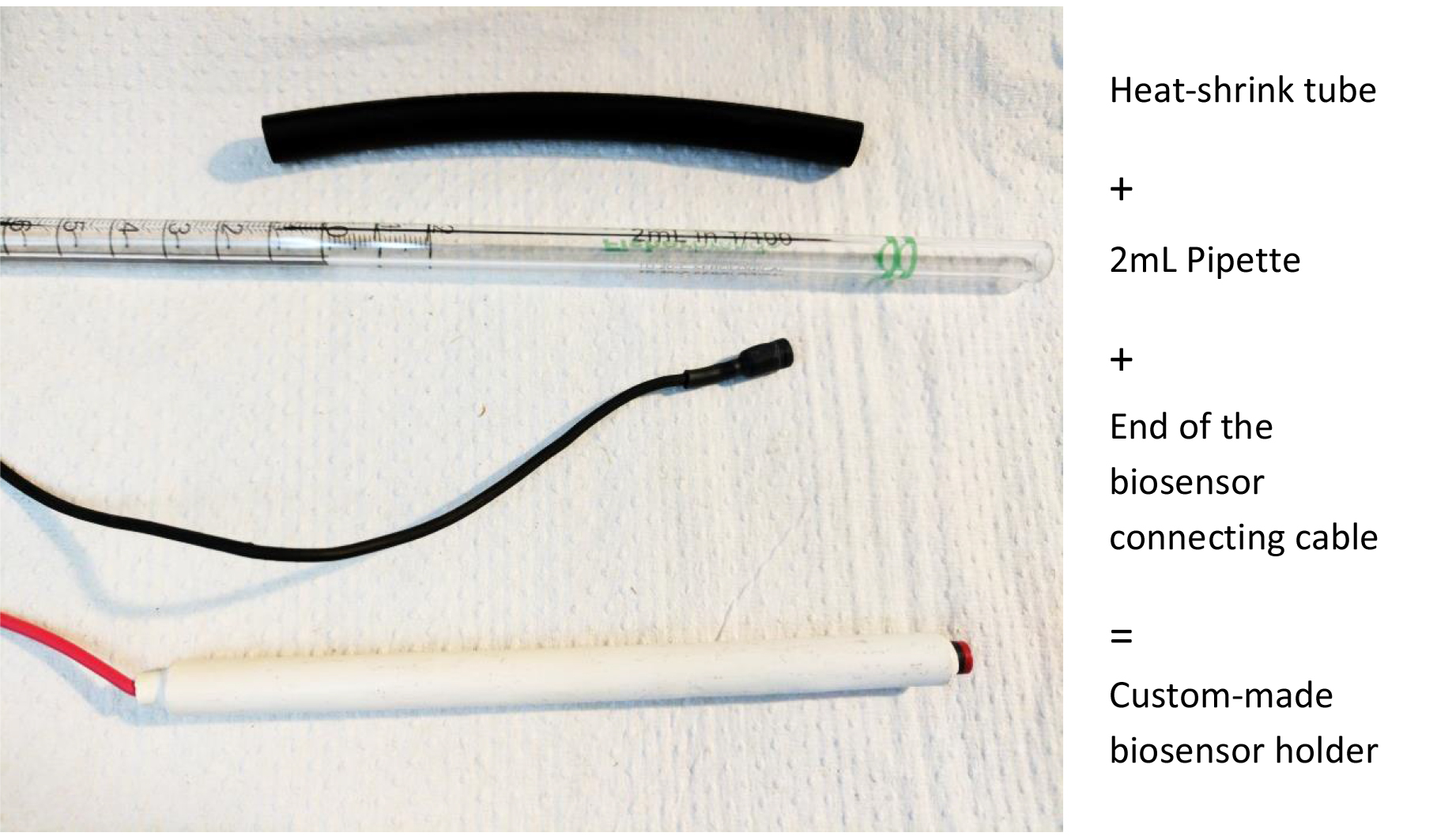
Figure 2. Making a biosensor holder. This will help manipulation and placement of the biosensors with manual manipulators.
- Chamber temperature controller (such as Warner Instruments, model: TC-344C , catalog number: 64-2401)
Software
- DY2000EN software (provided with bipotentiostat)
- Clampfit (version 9.2 minimum)
Note: If experiments are run on an electrophysiology set-up. Note that the DY2000EN software provides the sufficient analytical capabilities, but they were not used in our protocol.
Procedure
Before starting:
This protocol requires obtaining acute brain slices or acute slices from other tissue explants. Refer to Bio-protocol ‘Obtaining Acute Brain Slices’ (Papouin and Haydon, 2018) for a full and detailed description of that procedure. The present protocol requires the use of some material also described in Bio-protocol ‘Obtaining Acute Brain Slices’, such as a ‘nest beaker’ (Figure 3) and modified Pasteur pipette dropper (Figures 3 and 4). For a full description and instructions on how to build these, please refer to Bio-protocol ‘Obtaining Acute Brain Slices’.
1.Nest beaker
2.Modified Pasteur pipette dropper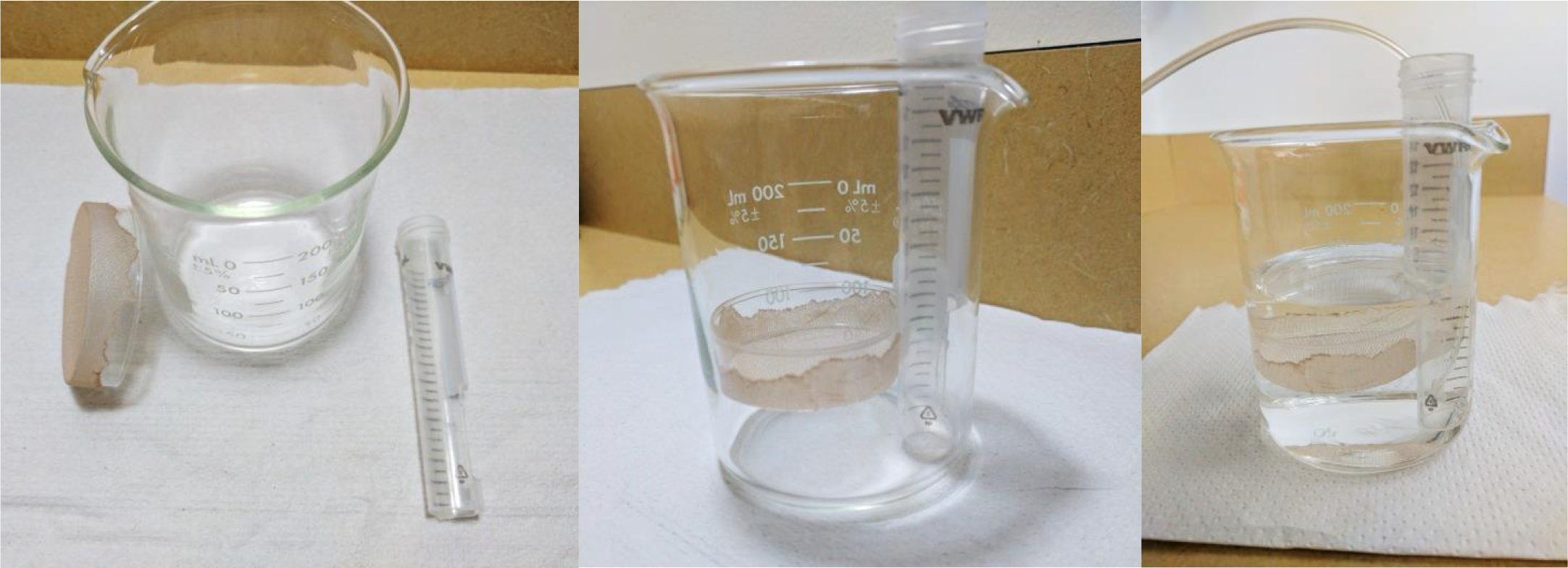
Figure 3. Nest beaker
Figure 4. Modifier Pasteur pipette dropper. Break the thinnest end of a glass disposable Pasteur pipet, and insert that end in a Dropper bulb.
Setting up:
- Prepare 1 L of stock ACSF the day prior and store overnight at 4 °C (see Recipes).
- On the day of the experiment, prepare 300 ml of ice-cold slicing ACSF (see Recipes) for acute brain slicing and refer to Bio-protocol ‘Obtaining Acute Brain Slices’ for a detailed procedure on how to obtain brain slices.
- Prepare 150 ml of recovery ACSF in the ‘nest beaker’ as instructed in Bio-protocol ‘Obtaining Acute Brain Slices’.
- Reserve the rest of the ACSF (~550 ml) to use as ‘Experimental ACSF’ (see Recipes)
- Obtain brain slices (or explants of the tissue of interest)
See Bio-protocol ‘Obtaining Acute Brain Slices’ Papouin and Haydon (2018). This is a very classic procedure used routinely in many labs for in vitro electrophysiology and various descriptions can be found in textbooks, on PubMed (for instance, Ting et al., 2014), or on Jove website (https://www.jove.com/video/2330). - Incubate slices to obtain conditioned medium
- In the case of the dorsal hippocampus, typically 6-8 hemi-slices should be obtained from an adult male mouse. This may vary depending on your brain region or tissue of interest. Once the recovery is over (Papouin and Haydon, 2018), transfer hemi-slices individually in a Petri dish filled with room temperature oxygenated recovery ACSF (from the nest beaker–a few ml will suffice), and carefully separate the hippocampus from the rest of the slice with spring scissors and fine forceps (Figure 5A). To minimize manipulation and damage to slices, a small portion of cortex immediately above the hippocampus (motor and somatosensory cortex typically) can be left attached (it would therefore participate in the following incubation and conditioned medium preparation). Transfer the ‘isolated’ hippocampal slices back to the nest beaker and allow an additional 15 min of recovery. This hippocampus isolation procedure can be greatly facilitated by removing the oxygenation tube from the Petri dish, on the condition that the procedure only takes 1 or 2 min and that slices are immediately transferred back to the nest beaker.
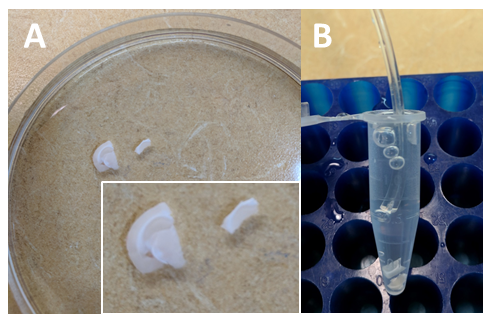
Figure 5. Incubation of hippocampal slices to obtain conditioned medium. A. In a Petri dish containing recovery ACSF, separate the hippocampus from the rest of the slice with spring scissors and fine forceps. B. After briefly transferring slices into experimental ACSF, transfer them into a 1.7 ml microtube filled to 1.5 ml with RT experimental ACSF. Oxygenate gently and leave in a tube rack for 90 min. - Using the modified Pasteur pipette (Figure 4), gently transfer all slices in a small Petri dish containing oxygenated experimental ACSF (see Recipes) at RT. Make sure the slices are not swirling around because of the oxygenation. This step is only required to transfer slices from recovery ACSF to conditioning ACSF (Step B3).
- After a few seconds, transfer 3 to 4 hippocampal slices at the bottom of a 1.7 ml microtube with the modified Pasteur pipette, and fill the tube up to 1.5 ml with RT experimental ACSF. Insert oxygenation tubing and gently oxygenate (Figure 5B). Reduce the rate of oxygenation so that the content of the tubes is not splashing and so that the slices are not swirling. Slices should remain at the bottom of the tube, gently waving. Incubate for 90 min at RT on a tube rack, while regularly checking that the oxygenation is appropriate.
- Carefully transfer the conditioned medium (CM) with a 1 ml pipette into a clean microtube and immediately freeze at -80 °C until used for D-serine measurement. If used at the same day, store at 4 °C. Be careful not to pipette any tissue with the CM. It is safer to leave a few µl in the tube. Unless they are treated differently, pool together the CM from all slice incubations in a 15 ml tube. The remaining experimental ACSF (i.e., vehicle ACSF) should be stored along with the CM samples, at -80 °C or 4 °C. Freeze the incubated tissue left at the bottom of the tubes at -80 °C until protein extraction and optic density measurement is performed with a Pierce BCA protein assay kit (not described in this protocol).
- In the case of the dorsal hippocampus, typically 6-8 hemi-slices should be obtained from an adult male mouse. This may vary depending on your brain region or tissue of interest. Once the recovery is over (Papouin and Haydon, 2018), transfer hemi-slices individually in a Petri dish filled with room temperature oxygenated recovery ACSF (from the nest beaker–a few ml will suffice), and carefully separate the hippocampus from the rest of the slice with spring scissors and fine forceps (Figure 5A). To minimize manipulation and damage to slices, a small portion of cortex immediately above the hippocampus (motor and somatosensory cortex typically) can be left attached (it would therefore participate in the following incubation and conditioned medium preparation). Transfer the ‘isolated’ hippocampal slices back to the nest beaker and allow an additional 15 min of recovery. This hippocampus isolation procedure can be greatly facilitated by removing the oxygenation tube from the Petri dish, on the condition that the procedure only takes 1 or 2 min and that slices are immediately transferred back to the nest beaker.
- Prepare biosensors
This protocol was designed for the use of Sarissa probes. It can easily be adapted for other D-serine sensors. If using Sarissa D-serine probes, prepare the biosensors according to manufacturer’s instructions Sarissa Biomedical (see also Dale et al., 2005) or as follows:- Run the vehicle ACSF (same ACSF that was used to obtain CM from the hippocampal slices) through the electrophysiology chamber and turn the chamber heater to 33 °C.
- Once the flow is steady and desired temperature is reached, mount the D-serine and the Null sensor probes on the biosensor-holder attached to the micromanipulators (Figure 1).
- Using the micromanipulators, submerge the probes in the recording chamber and position them so that they are close to each other and at the same depth (Figures 1 and 6). Preferably, the two probes should be facing each other, at a similar angle respective to the flow. Let sensors rehydrate for 15-30 min. Note that while rehydrating, the sensors will swell in size (like a sponge). Give the sensors enough space to rehydrate without coming into contact with one another or the bottom of the chamber. The probe part of the biosensors is extremely fragile: make sure it does not come into contact with anything that could damage it.
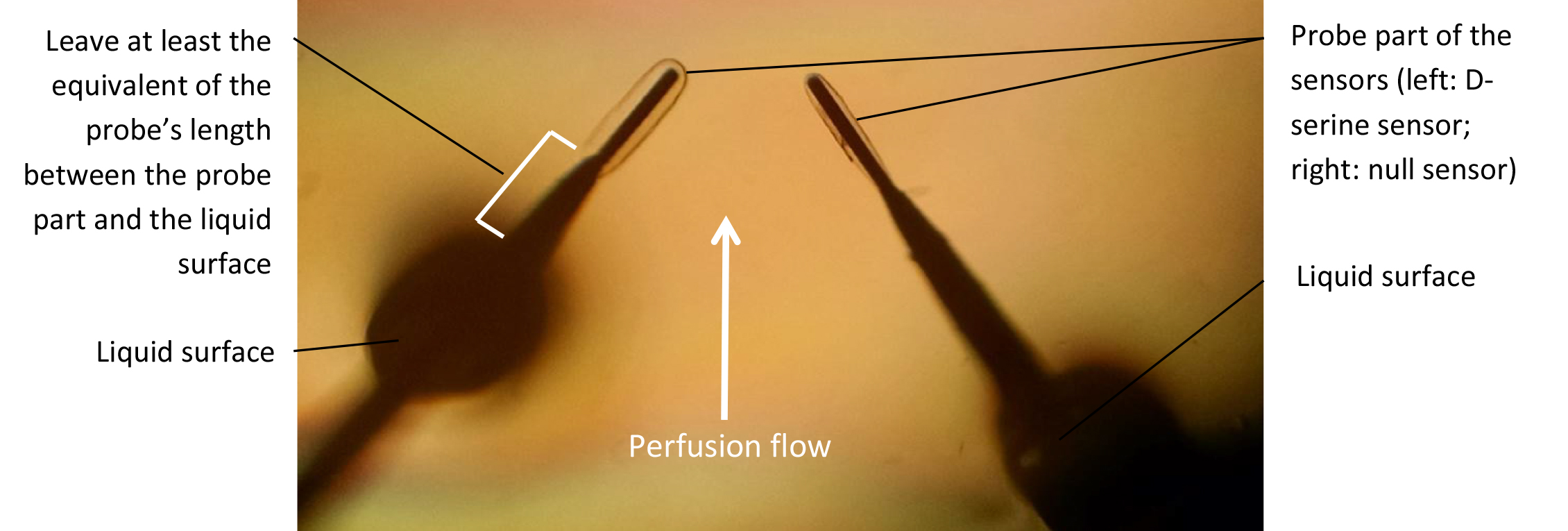
Figure 6. Biosensor probe submersion and positioning in the recording chamber
Notes: Before starting ‘D- Run conditioned medium on biosensors’ make sure the perfusion line is as short as possible. Make sure the perfusion rate is no greater than 0.5 ml/min. Make sure that the sensor part of the probe is constantly submerged. Once the probes are rehydrated, they cannot be exposed to the air more than a few seconds. Exposing the sensors to the air while polarized at +500 mV will result in immediate and irreversible damage to the sensor. We strongly recommend running a trial run with dummy sensors. - Using the bipotentiostat and the polarity command on the software interface, manually polarize the sensors at +500 mV and switch the polarity to -500 mV and back, cycling back and forth about 10 to 12 times (see Figure 7).
- Allow polarizing to +500 mV for at least 60 min or until capacitive decay is imperceptible over the course of a 15-20 min ‘run’ (Figure 7).
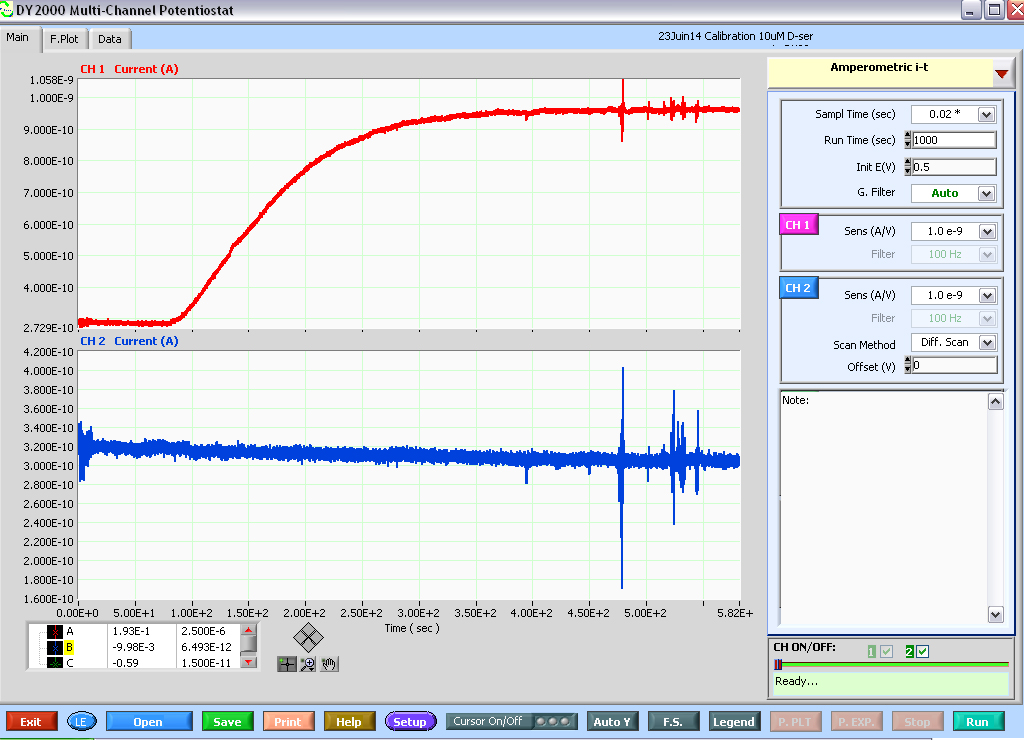
Figure 7. Print screen of a calibration experiment (10 μM D-serine) run on a D-serine sensor (top red trace) and null sensor (bottom blue trace) using the DY2000 software. Note the parameters used, in particular, sampling rate: 50 Hz, Duration of a run: 1,000 sec, Sensors potential: 0.5 V and A/V scale factors. Note that at the end of each run, the software will show a ‘Ready…’ signal (bottom right-hand corner), signaling that a new run can be started (by pressing the RUN button, bottom right-hand corner). However, the run that just ended is not automatically saved. Make sure to press Save (bottom left, green button) prior to starting another run or data will be lost. Also note that ‘noise’ artifacts typically appear on both traces. However, D-serine perfusion elicits no current on the null sensor. In fact, one can still detect a ~20 pA capacitive decay over the course of 1,000 sec recording on the null sensor trace.
- Run the vehicle ACSF (same ACSF that was used to obtain CM from the hippocampal slices) through the electrophysiology chamber and turn the chamber heater to 33 °C.
- Run conditioned medium on biosensors
- Thaw the conditioned media on ice and then leave at RT. Tubes may contain some deposit. Do not perfuse the deposit through the line, instead pipette out the CM into a clean tube. Quickly switch the perfusion line from the vehicle solution to the tube containing the CM. Make sure there are no bubbles in the line (we recommend ‘pinching’ the perfusion line when switching solutions).
- Once the volume of CM runs through, switch the line back to the vehicle solution for as long as necessary to obtain a full wash-in/wash-out curve (Figure 8) and to clean the line of residual D-serine before the next measurement.
- If the next CMs to be run on the sensors were obtained on a different day using different vehicle ACSF, run the corresponding vehicle solution for at least 15 min before running the next CM. Make sure the sensors are not exposed to the air during this process.
Note: The DY2000 software provided with the biopotentiostat requires saving the results from each run after the run is complete. It is therefore critical to remember to click SAVE at the end of each induvial run (Figure 7). - It appears that different sensors tend to give very different reading qualities and steadiness. We recommend proceeding to Procedure D only once, it is ascertained that the set of biosensors provides a steady and clean amperometric signal devoid of artifacts and fluctuations due to perfusion flow or electrical noise. In our study, we chose to abort or exclude all experiments in which the baseline behavior of the sensors would prevent accurate and reliable evaluation of D-serine levels in CM.
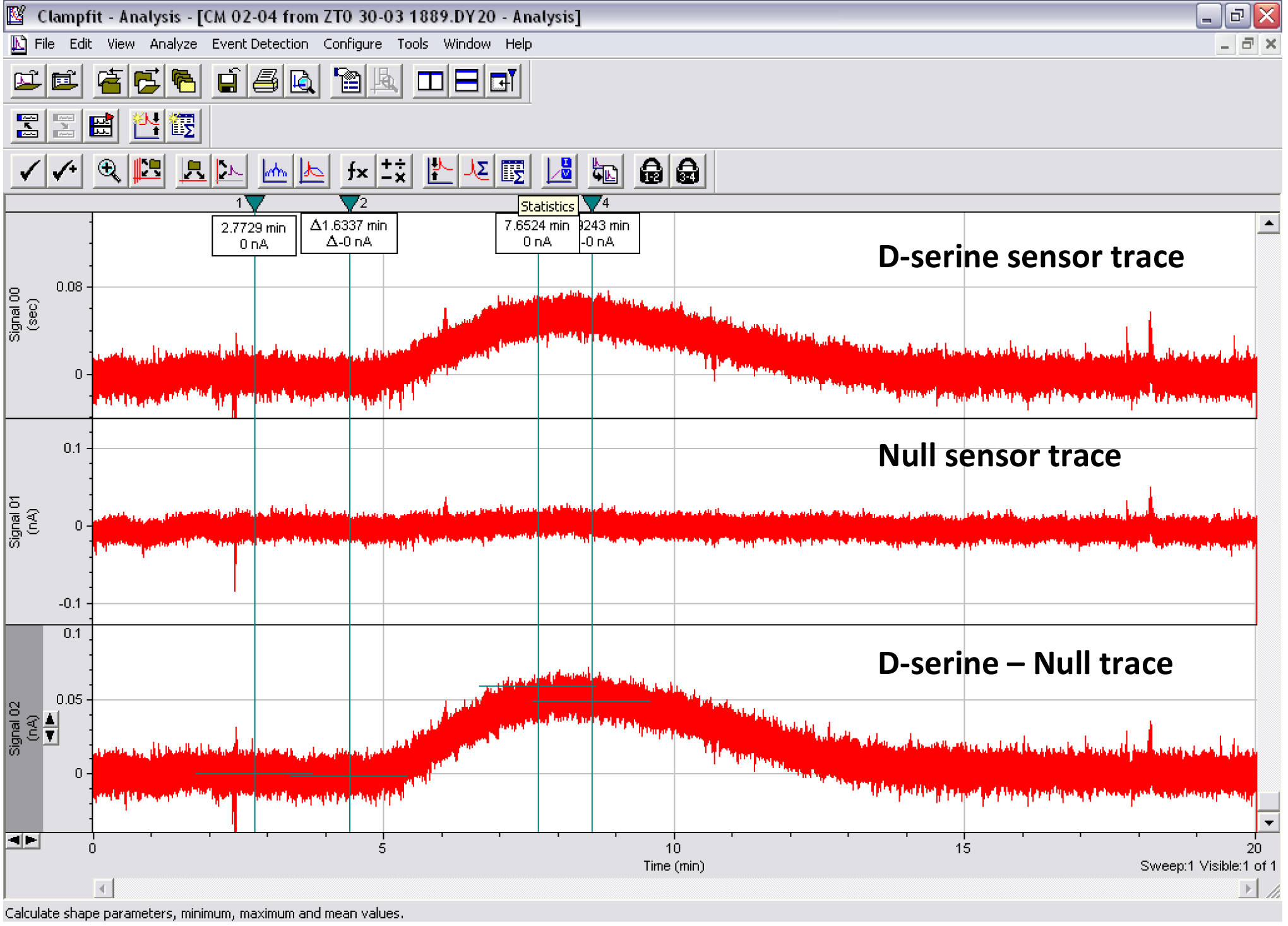
Figure 8. Print screen of the Clampfit analysis of a CM D-serine measurement. Top trace: D-serine sensor. Middle trace: Null sensor. Bottom trace: D-serine sensor minus null sensor. Cursors 1-2 show where the baseline subtraction is made so that the baseline portion of the trace reads 0 pA. Cursors 3-4 show where the mean peak response is measured.
- Thaw the conditioned media on ice and then leave at RT. Tubes may contain some deposit. Do not perfuse the deposit through the line, instead pipette out the CM into a clean tube. Quickly switch the perfusion line from the vehicle solution to the tube containing the CM. Make sure there are no bubbles in the line (we recommend ‘pinching’ the perfusion line when switching solutions).
- Calibrate the biosensors
- In advance, prepare standard solutions of D-serine of, for example, 0.05, 0.5, 5 and 50 µM using serial dilutions in vehicle ACSF. We recommend increasing the accuracy of the calibration around the concentrations of D-serine you expect to detect in the CM.
- Switch from the vehicle solution to the D-serine standard solution, beginning with the lowest concentration standard.
- Run each of the D-serine standard solutions through the line until a plateau is reached (Figure 9) before switching back to vehicle ACSF. Allow sufficient time between each solution to thoroughly and completely wash-out after each application (this can take up to 20 min for the highest concentrations depending on your set-up).
Notes: For Procedures D and E, when switching from one solution to the other, make sure no interruption in the perfusion flow occurs to guarantee that the amount of liquid (and liquid flow) remains constant in the chamber. Proceeding otherwise could compromise the immersion of the sensors and expose and irreversibly damage them.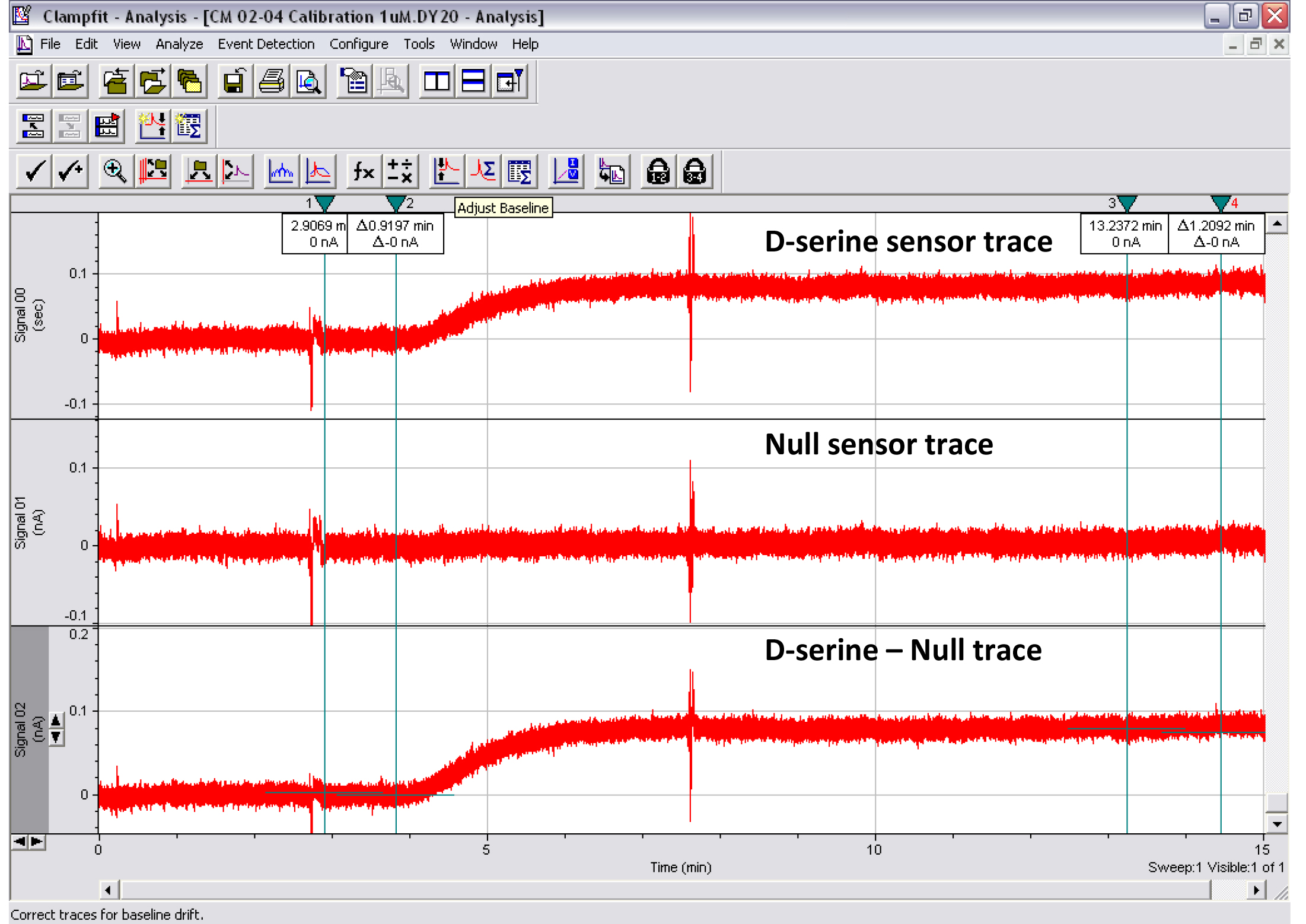
Figure 9. Print screen of the Clampfit analysis of a 1 μM D-serine calibration experiment. Top trace: D-serine sensor. Middle trace: Null sensor. Bottom trace: D-serine sensor minus null sensor. Cursors 1-2 show where the baseline subtraction is made so that the baseline trace reads 0 pA. Cursors 3-4 show where the mean peak response is measured. Artifacts were intentionally created on traces around the start of perfusion of the 1 μM D-serine solution and after ~3 ml of perfusion. This type of experiment was used to determine the minimal volume of liquid required to measure D-serine concentration in CM reliably.
- In advance, prepare standard solutions of D-serine of, for example, 0.05, 0.5, 5 and 50 µM using serial dilutions in vehicle ACSF. We recommend increasing the accuracy of the calibration around the concentrations of D-serine you expect to detect in the CM.
- Discard or store the biosensors
- Make sure that the biosensors are no longer polarized and the bipotentiostat is turned off before handling and unplugging the biosensors.
- If sensors are to be reused, follow manufacturer’s instructions. Quickly transfer the sensors to Buffer A (see manufacturer’s instructions) in a rehydration pot at 4 °C and place in a fridge. Alternatively, from our experience, we found that if handled properly, sensors can be kept overnight in cold ACSF (4 °C) and reused at least once within the next 48 h without major loss of sensitivity.
- Make sure that the biosensors are no longer polarized and the bipotentiostat is turned off before handling and unplugging the biosensors.
Data analysis
The data analysis can be run using Clampfit, like any electrophysiology trace (see Figures 8 and 9). However, this requires exporting the data as an Excel file beforehand. Follow these quick simple steps to do so.
- Right-click on the file to be analyzed.
- Open with Excel.
- Delete the first rows that contain details about the experiment.
- Optional: Delete the time column if you know the sampling rate. Add an additional column that corresponds to the signal from the null sensors subtracted from that of the D-serine sensor (D-ser–Null sensor). Note that this subtraction can also be done on Clampfit.
- Save as a separate text file (.txt).
- Open that text file with Clampfit.
Note: Clampfit will ask the number of signals, sampling rate, units, scale, and time frame. Give pertinent information with regard to point #4 above. - To obtain the D-serine–Null sensor trace, if this was not done on the Excel text file, save each trace individually (as a D-serine and a Null trace) and, while both files are open simply open the tab Analyze > subtract control. The D-serine–Null sensor trace will appear as a separate individual trace and can be saved as a separate file.
- Using Clampfit cursors and tools, zero the trace at baseline and measure the peak response of the D-serine–Null sensor trace. We recommend measuring the average current value (in pA) over one or several minutes of the plateau response for more accuracy, instead of simply measuring the ‘absolute peak’ which might give false values given that traces are usually noisy (Figures 8 and 9).
- D-serine biosensors have a specified lower detection limit of 100 nM. However, in our hands, they successfully detected concentrations as low as 50 nM (when running D-serine standards). It is up to the experimenter’s discretion to decide whether D-serine measurements falling below the specified detection limit should be excluded or whether they should be considered accurate. In our study (Papouin et al., 2017b), we chose to be conservative and show such data points but exclude them from the overall analysis.
Notes
- The volume of CM incubated with slices (3 ml in this case) should be set by preliminary experiments aimed at determining the minimal volume of CM that can be run through the perfusion system that allows plateau detection by the biosensors (see Figure 9 and Legend).
- Though no current is generally observed on the Null sensor, currents measured on this sensor electrode must be subtracted from currents measured on the D-serine sensor electrode to obtain a pure ‘D-serine-induced’ amperometric signal; or ‘net D-serine signal’.
- Sensors should be calibrated at the end of every experiment to measure their sensitivity. For the first few experiments, we recommend calibrating the sensors before running the CM. This will allow inexperienced experimenters to familiarize themselves with the procedure. This also presents the advantage that if any problem were to occur over the course of the experiments (such as a perfusion issue and damage to the biosensors), it will still be possible to determine the absolute concentration of D-serine in all CM run until then. However, it is unknown whether running high concentrations of D-serine (50-100 µM) for several minutes at a time causes any loss of DAAO activity or integrity, and whether, therefore, this causes a loss of sensitivity of the sensors. Since D-serine levels in the CM are typically low, this could result in a lack of detection or underestimation in the following D-serine measurements. One also runs the risk of residual subthreshold amounts of D-serine being present in the line or chamber and false detection during the subsequent CM perfusion.
- Classical use of these biosensors normally consists of penetrating brain slices with the biosensor electrode themselves (Shigetomi et al., 2013). We strongly advise against using this method as it will inevitably result in an immediate peak detection of tens of mM of D-serine, due to tissue destruction and irreversible damage to cells, causing intracellular D-serine (which accounts for 99.9% of brain D-serine) to be released into the extracellular space. After tens of minutes of decay, this is expected to stabilize to a steady-state level thought to reflect the endogenous extracellular concentration of D-serine in slices. Instead, this is more likely to reflect residual amounts of D-serine escaping the scarring/dying tissue surrounding the biosensor. Additionally, this approach is highly dependent on how deep sensors are forced into the slice, which dictates how much of the sensor is in direct contact with damaged tissue and therefore how much D-serine is detected. Last but not least, we found that such a method could cause deterioration of the sensor itself upon penetration in the slice. Together these concerns encouraged us to employ a different approach described in this protocol.
Recipes
- Stock artificial cerebrospinal fluid (ACSF) solution (1 L, store at 4 °C)
Glucose 10 mM (1.8 g for 1 L)
Sodium chloride 125 mM (7 g for 1 L)
Sodium phosphate monobasic anhydrous 1.25 mM (0.119 g for 1 L)
Potassium chloride 2.5 mM (0.23 mg for 1 L)
Sodium bicarbonate 26 mM (2.18 mg for 1 L)
Adjust pH 7.3 and osmolarity 290-300 mOsm L-1
Make up to 1 L with ddH2O - Ice-cold slicing ACSF (~300 ml)
Stock ACSF
2 mM magnesium chloride
1 mM calcium chloride - Recovery ACSF (~150 ml)
Stock ACSF
1.5 mM magnesium chloride
2 mM calcium chloride - Experimental ACSF (~550 ml)
Stock ACSF
1.3 mM magnesium chloride
2 mM calcium chloride
Note: Of which ~3 ml will be used to obtained hippocampal slices-conditioned medium (conditioning ACSF) and the rest will be used as a ‘blank’ during the amperometric D-serine measurements (vehicle ACSF). Label and store at RT for < 2 days or freeze at -20 °C. Each conditioned medium must have its corresponding vehicle ACSF.
Acknowledgments
This work was supported by two Philippe Foundation grants and a Human Frontier Science Program long-term fellowship (LT000010/2013) awarded to T.P., and two NIH/NINDS R01 grants (NS037585 and AA020183) awarded to P.G.H. who is also the founder of GliaCure. Authors declare no conflict of interest. We thank Jaclyn M. Dunphy for her careful proofreading of this protocol.
References
- Dale, N., Hatz, S., Tian, F. and Llaudet, E. (2005). Listening to the brain: microelectrode biosensors for neurochemicals. Trends Biotechnol 23(8): 420-428.
- Ferreira, J. S., Papouin, T., Ladepeche, L., Yao, A., Langlais, V. C., Bouchet, D., Dulong, J., Mothet, J. P., Sacchi, S., Pollegioni, L., Paoletti, P., Oliet, S. H. R. and Groc, L. (2017). Co-agonists differentially tune GluN2B-NMDA receptor trafficking at hippocampal synapses. Elife 6.
- Johnson, J. W. and Ascher, P. (1987). Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature 325(6104): 529-531.
- Kleckner, N. W. and Dingledine, R. (1988). Requirement for glycine in activation of NMDA-receptors expressed in Xenopus oocytes. Science 241(4867): 835-837.
- Montesinos Guevara, C. and Mani, A. R. (2016). The role of D-serine in peripheral tissues. Eur J Pharmacol 780: 216-223.
- Papouin, T., Dunphy, J. M., Tolman, M., Dineley, K. T. and Haydon, P. G. (2017a). Septal cholinergic neuromodulation tunes the astrocyte-dependent gating of hippocampal NMDA receptors to wakefulness. Neuron 94(4): 840-854 e847.
- Papouin, T and Haydon, P. G. (2018). Obtaining acute brain slices. Bio-protocol 8(2): e2699.
- Papouin, T., Henneberger, C., Rusakov, D. A. and Oliet, S. H. R. (2017b). Astroglial versus neuronal D-serine: Fact checking. Trends Neurosci 40(9): 517-520.
- Papouin, T., Ladepeche, L., Ruel, J., Sacchi, S., Labasque, M., Hanini, M., Groc, L., Pollegioni, L., Mothet, J. P. and Oliet, S. H. (2012). Synaptic and extrasynaptic NMDA receptors are gated by different endogenous coagonists. Cell 150(3): 633-646.
- Pernot, P., Mothet, J. P., Schuvailo, O., Soldatkin, A., Pollegioni, L., Pilone, M., Adeline, M. T., Cespuglio, R. and Marinesco, S. (2008). Characterization of a yeast D-amino acid oxidase microbiosensor for D-serine detection in the central nervous system. Anal Chem 80(5): 1589-1597.
- Shigetomi, E., Jackson-Weaver, O., Huckstepp, R.T., O'Dell, T.J., Khakh, B.S. (2013). TRPA1 channels are regulators of astrocyte basal calcium levels and long-term potentiation via constitutive D-serine release. J Neurosci 33(24): 10143-53.
- Ting, J.T., Daigle, T.L., Chen, Q., Feng, G. (2014). Acute brain slice methods for adult and aging animals: application of targeted patch clamp analysis and optogenetics. Methods Mol Biol 1183: 221-42.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Papouin, T. and Haydon, P. G. (2018). D-serine Measurements in Brain Slices or Other Tissue Explants. Bio-protocol 8(2): e2698. DOI: 10.21769/BioProtoc.2698.
Category
Neuroscience > Cellular mechanisms > Synaptic physiology
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









