- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro RNA-dependent RNA Polymerase Assay Using Arabidopsis RDR6
Published: Vol 8, Iss 1, Jan 5, 2018 DOI: 10.21769/BioProtoc.2673 Views: 10933
Reviewed by: Tie LiuLaia ArmengotPooja Verma

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1465 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3196 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1812 Views
Abstract
RNA-dependent RNA polymerases (RdRPs) in eukaryotes convert single-stranded RNAs into double-stranded RNAs, thereby amplifying small interfering RNAs that play crucial roles in the regulation of development, maintenance of genome integrity and antiviral immunity. Here, we describe a method of in vitro RdRP assay using recombinant Arabidopsis RDR6 prepared by an insect expression system. By using this classical biochemical assay, we revealed that RDR6 has a strong template preference for RNAs lacking a poly(A) tail. This simple method will be applicable to other RdRPs in Arabidopsis and different organisms.
Keywords: RNA-dependent RNA polymeraseBackground
RNA-dependent RNA polymerase (RdRP) genes have been found in all eukaryotic kingdoms–plants, fungi, protista and animals (Zong et al., 2009). They convert single-stranded RNAs (ssRNAs) into double-stranded RNAs (dsRNAs), thereby amplifying small interfering RNAs (siRNAs) that play crucial roles in various biological processes including regulation of development (Peragine et al., 2004; Li et al., 2005), maintenance of genome integrity (Volpe et al., 2002; Xie et al., 2004) and antiviral immunity (Mourrain et al., 2000; Yu et al., 2003; Garcia-Ruiz et al., 2010; Wang et al., 2010). In addition to this RdRP activity, RdRPs possess another enzymatic activity called terminal nucleotide transferase (TNTase) activity (Curaba and Chen, 2008; Aalto et al., 2010), which adds one or more nucleotides to the 3’ end of ssRNAs or dsRNAs in a template-independent manner.
Here, we describe a method for in vitro RdRP assay using recombinant Arabidopsis thaliana RDR6 prepared by an insect expression system. To accurately discriminate RdRP products from TNTase products, we designed two strategies: 1) elimination of single-stranded TNTase products by treating the reaction mixture with ssRNA-specific RNase I, and 2) electrophoresis of the reaction mixture on a native acrylamide gel, in which double-stranded RdRP products can be distinguished from single-stranded TNTase products based on their different mobility. By using this classical biochemical assay, we revealed that RDR6 has a strong template preference for the RNAs lacking a poly(A) tail (Baeg et al., 2017). This simple method should also be applied to other RdRPs in Arabidopsis and different organisms.
Materials and Reagents
- Preparation of recombinant RDR6
- 100 mm dish (Corning, catalog number: 430167 )
- 50 ml conical centrifuge tube (Nippon Genetics, catalog number: FG200 )
- Cell counter plate (Neubauer Improved) (FUKAEKASEI and WATSON, WATSON BIOLAB, catalog number: 177-112C )
- 1.5 ml tube (BM Equipment, BMBio, catalog number: NT-175 )
- Drosophila melanogaster Schneider 2 (S2) cells (Schneider, 1972)
- RDR6 expression plasmid for S2 cells (pAFW-SUMO-AtRDR6 [Baeg et al., 2017])
- Anti-FLAG antibody, M2 (Sigma-Aldrich, catalog number: F1804 )
- SUMOstar protease (LifeSensors, catalog number: 4110 )
- Schneider’s Drosophila medium (Thermo Fisher Scientific, GibcoTM, catalog number: 21720001 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 12483020 )
- Antibiotic-Antimycotic (100x) (Thermo Fisher Scientific, GibcoTM, catalog number: 15240062 )
- X-tremeGENE HP transfection reagent (Sigma-Aldrich, Roche Diagnostics, catalog number: 06366546001 )
- Dynabeads proteins G (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10009D )
- Triton X-100 (Wako Chemical Pure Industries, catalog number: 169-21105 )
- Dithiothreitol (DTT) (NACALAI TESQUE, catalog number: 14128-04 )
- Glycerol (NACALAI TESQUE, catalog number: 17018-83 )
- Liquid nitrogen
- PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 23227 )
- Sodium chloride (NaCl) (Wako Chemical Pure Industries, catalog number: 190-13921 )
- Potassium chloride (KCl) (NACALAI TESQUE, catalog number: 28514-75 )
- Sodium phosphate dibasic dodecahydrate (Na2HPO4·12H2O) (Sigma-Aldrich, catalog number: 28-3720 )
- Potassium dihydrogenphosphate (KH2PO4) (NACALAI TESQUE, catalog number: 28721-55 )
- HEPES (DOJINDO, catalog number: 340-01376 )
- Magnesium acetate tetrahydrate, Mg(OAc)2 (Wako Chemical Pure Industries, catalog number: 130-00095 )
- EDTA-free protease inhibitor cocktail (Sigma-Aldrich, catalog number: 11836170001 )
- Potassium acetate (KOAc) (NACALAI TESQUE, catalog number: 28405-05 )
- Phosphate-buffered saline (PBS) (see Recipes)
- Hypotonic lysis buffer (see Recipes)
- 1x lysis buffer (see Recipes)
- Preparation of template RNAs
- Fluorescent TLC plate
- 2 ml tube (BM Equipment, BMBio, catalog number: BM-20 )
- Sterilized pipette tips
- Plastic wrap (AsahiKASEI)
- T7-ScribeTM Standard RNA IVT Kit (CELLSCRIPT, catalog number: C-AS3107 )
- Distilled water
- Phenol:chloroform:isoamyl alcohol 25:24:1 mixed, pH 5.2 (NACALAI TESQUE, catalog number: 26058-96 )
- Chloroform (Wako Chemical Pure Industries, catalog number: 035-02616 )
- Isoamyl alcohol (Wako Chemical Pure Industries, catalog number: 135-12015 )
- Ammonium acetate (NACALAI TESQUE, catalog number: 02433-35 )
- 70% ethanol
- 2-Propanol (NACALAI TESQUE, catalog number: 29113-53 )
- Ethylenediaminetetraacetic acid (EDTA) (NACALAI TESQUE, catalog number: 15105-35 )
- Deionized formamide (NACALAI TESQUE, catalog number: 16229-95 )
- Xylene cyanol (Wako Chemical Pure Industries, catalog number: 240-00463 )
- Tris (Wako Chemical Pure Industries, catalog number: 207-06275 )
- Sodium chloride (NaCl) (Wako Chemical Pure Industries, catalog number: 190-13921 )
- Sodium dodecyl sulfate (SDS) (NACALAI TESQUE, catalog number: 02873-75 )
- Bromophenol blue (Wako Chemical Pure Industries, catalog number: 029-02912 )
- Urea (Wako Chemical Pure Industries, catalog number: 217-00171 )
- SequaGel-UreaGel Concentrate (National Diagnostics, catalog number: EC-830 )
- 2x formamide dye (see Recipes)
- PK buffer (see Recipes)
- Fluorescent TLC plate
- RNA-dependent RNA polymerase activity assay
- RNasin® Plus RNase Inhibitor (Promega, catalog number: N2615 )
- α-32P-UTP (PerkinElmer, catalog number: NEG507H )
- 100% ethanol (NACALAI TESQUE, catalog number: 14713-53 )
- Ribonucleotide Solution set (New England Biolabs, catalog number: N0450L )
- Glycogen (NACALAI TESQUE, catalog number: 17110-11 )
- Proteinase K (20 mg/ml) (NACALAI TESQUE, catalog number: 29442-85 )
- Boric acid (H3BO3) (NACALAI TESQUE, catalog number: 05215-05 )
- Ethylenediaminetetraacetic acid (EDTA) (NACALAI TESQUE, catalog number: 15105-35 )
- HEPES (DOJINDO, catalog number: 340-01376 )
- Ammonium acetate (NH4OAc) (NACALAI TESQUE, catalog number: 02433-35 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Wako Chemical Pure Industries, catalog number: 135-00165 )
- RNase I (Thermo Fisher Scientific, InvitrogenTM, catalog number: AM2294 )
- PEG 4000 (Sigma-Aldrich, catalog number: 95904-250G-F )
- Glycerol (NACALAI TESQUE, catalog number: 17018-83 )
- Tartrazine (NACALAI TESQUE, catalog number: 32706-22 )
- 40(w/v)%-Acrylamide/Bis Mixed Solution(19:1) (NACALAI TESQUE, catalog number: 06140-45 )
- 1 mM NTP mixture (see Recipes)
- PK buffer (EDTA-) (see Recipes)
- PK mixture (see Recipes)
- PK mixture (EDTA-) (see Recipes)
- 5x TBE (see Recipes)
- 10x RdRP buffer (see Recipes)
- 2x RNase I buffer (see Recipes)
- 2x loading buffer (see Recipes)
- RNasin® Plus RNase Inhibitor (Promega, catalog number: N2615 )
Equipment
- Pipettes (Gilson)
- Cell incubator (SANYO, catalog number: MIR-153 )
- Centrifuge (TOMY SEKIO, catalog number: MX-301 )
- Vortex mixer (Scientific Industries, model: Vortex-Genie 2 , catalog number: SI-0286)
- Rotator (TAITEC, model: RT-30mini, catalog number: 0057154-000 )
- Magnetic stand (Thermo Fisher Scientific, catalog number: 120.20 )
- Block incubator (TAITEC)
- UV lamp (UVP, catalog number: 95-0004-11 )
- Gel dryer (Bio-Rad Laboratories, catalog number: 165-1745 )
- Imaging plate cassette (GE Healthcare, catalog number: 63-0035-45 )
- Imaging plate (GE Healthcare, catalog number: 28-9564-74 )
- Typhoon FLA 7000 IP (GE Healthcare, model: Typhoon FLA 7000 IP )
Procedure
- Preparation of recombinant RDR6
The recombinant RDR6 protein with N-terminal 3xFLAG-tags followed by a SUMOstar protease cleavage site is overexpressed by using Drosophila S2 cell expression system. This system produces higher levels of soluble recombinant RDR6 compared to plant cell-free translation systems or Escherichia coli expression systems. After lysis of the cells, the recombinant protein is purified by anti-FLAG antibody conjugated magnetic beads, and eluted with SUMOstar protease. Although we use the tag-less recombinant RDR6 protein, the N- or C-terminally tagged RDR6 can also be used for RdRP activity assay (Devert et al., 2015; Baeg et al., 2017).- When S2 cells reach between 1.0 x 107 and 1.5 x 107 cells/ml in a 100-mm dish, collect them into a 50-ml tube(s). Centrifuge at 1,500 x g for 3 min, remove supernatant, and resuspend the cell pellet gently with Schneider’s Drosophila medium containing 10% FBS. Count the cell numbers using a cell counter plate. Seed 1.5 x 107 cells (1.5 x 106 cells/ml) in a 100-mm dish in 10 ml of Schneider’s Drosophila medium containing 10% FBS (see Notes 1 and 2).
- Dilute 10 μl of 1 μg/μl pAFW-SUMO-AtRDR6 plasmid with 500 μl of Schneider’s Drosophila medium in a 1.5 ml tube. Add 20 μl of X-tremeGENE HP transfection reagent to the diluent, mix by tapping, and incubate it at room temperature (~25 °C) for 10 min.
- Add the transfection mixture into S2 cells in a dropwise manner, gently agitate the dish in a circular motion to evenly disperse the transfection mixture, and statically incubate at 27 °C for 72 h.
- Collect the cells into a 50-ml conical centrifuge tube(s). Centrifuge in a swinging bucket rotor at 1,500 x g for 3 min at 25 °C. Remove the supernatant with a pipette.
- Resuspend the cell pellet gently in 300 μl of PBS (see Recipes). Transfer the cell suspension into a 1.5 ml tube. Centrifuge at 1,500 x g for 3 min at 4 °C. Remove the supernatant with a pipette completely.
- Weigh the cells and add an equal amount of pre-chilled hypotonic lysis buffer (see Recipes). Resuspend the cell pellet gently by tapping the bottom of the tube.
- Incubate the cell suspension on ice for 15 min, then vortex the cell suspension for 30 sec to disrupt the cell membrane.
- Centrifuge at 17,000 x g for 20 min at 4 °C.
- Transfer the supernatant into the 1.5 ml tube containing Dynabeads protein G coated with anti-FLAG antibody (see Note 3). Mix by tapping, and incubate on a rotator at 4 °C for 1 h.
- Transfer the tube to a magnetic stand. Wash the beads twice with an equal volume of 1x lysis buffer (see Recipes) containing 800 mM NaCl and 1% Triton X-100, and once with an equal volume of 1x lysis buffer.
- Resuspend the beads with 100 μl of 1x lysis buffer containing 1 mM DTT, 20% glycerol and 0.05 U per μl SUMOstar protease. Incubate the slurry on a rotator at 4 °C for 20 min.
- Recover the supernatant in a pre-chilled tube, aliquot it into several tubes on ice, freeze with liquid nitrogen, and store at -80 °C. Measure the concentration of purified RDR6 by SDS-PAGE along with the defined dilutions of bovine serum albumin standard (PierceTM BCA Protein Assay Kit) after Coomassie Brilliant Blue (CBB) staining (Figure 1).
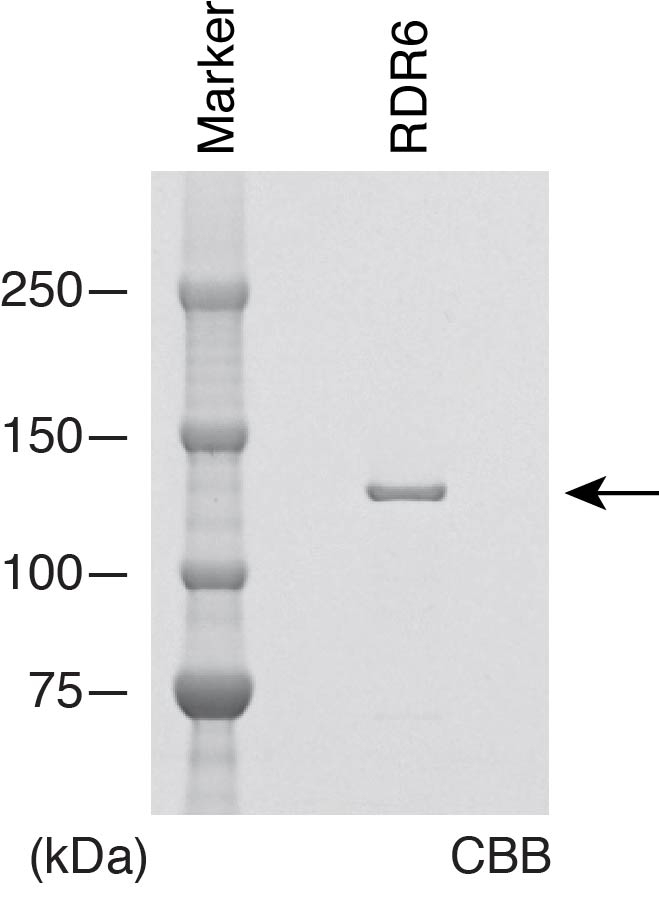
Figure 1. Recombinant RDR6. CBB staining of purified recombinant protein. The arrow indicates purified recombinant RDR6.
- When S2 cells reach between 1.0 x 107 and 1.5 x 107 cells/ml in a 100-mm dish, collect them into a 50-ml tube(s). Centrifuge at 1,500 x g for 3 min, remove supernatant, and resuspend the cell pellet gently with Schneider’s Drosophila medium containing 10% FBS. Count the cell numbers using a cell counter plate. Seed 1.5 x 107 cells (1.5 x 106 cells/ml) in a 100-mm dish in 10 ml of Schneider’s Drosophila medium containing 10% FBS (see Notes 1 and 2).
- Preparation of template RNAs
Because in vitro transcribed RNAs often contain unwanted shorter or longer byproducts, polyacrylamide gel electrophoresis (PAGE) purification of RNAs is essential for accurate determination of the template specificity of RdRPs. When synthetic oligo RNAs are used as the template for RdRPs, the use of HPLC purification is recommended.- Transcribe reporter RNAs from DNA templates at 37 °C for 2 h using T7-Scribe Standard RNA IVT Kit following manufacturer’s instructions.
- Add 1 μl of DNase I, mix thoroughly and incubate at 37 °C for 20 min.
- Adjust the volume of the reaction mixture to 100 μl with distilled water on ice.
- Add an equal volume of phenol:chloroform:isoamyl alcohol into the reaction mixture, and mix by vortexing at room temperature (~25 °C). Centrifuge at 17,000 x g for 2 min at 4 °C.
- Recover the aqueous phase into a new tube with an equal volume of chloroform:isoamyl alcohol, and mix by vortexing at room temperature (~25 °C). Centrifuge at 17,000 x g for 1 min at 4 °C.
- Recover the aqueous phase into a new chilled tube, add an equal volume of pre-chilled 5 M ammonium acetate, mix the solution by tapping the tube, and incubate on ice for 15 min.
- Centrifuge at 21,500 x g for 20 min at 4 °C.
- Remove the supernatant and add 700 μl of 70% ethanol.
- Centrifuge at 17,000 x g at 4 °C for 3 min and remove the supernatant completely.
- Place the open tube on ice to air dry.
- Resuspend the pellet in 40 μl of 1x formamide dye (see Recipes). Incubate the sample at 95 °C for 5 min, mix by vortexing and load onto a urea gel containing 0.5x TBE and appropriate concentration of polyacrylamide (see Notes 4 and 5).
- Perform electrophoresis (see Note 6).
- Transfer the gel onto a plastic wrap, cover the gel with another wrap, and place it on a fluorescent TLC plate. Briefly expose the gel to UV light and excise a gel slice containing the RNA of interest (see Note 7). Transfer the gel slice into a 2 ml tube (Figures 2A-2C).
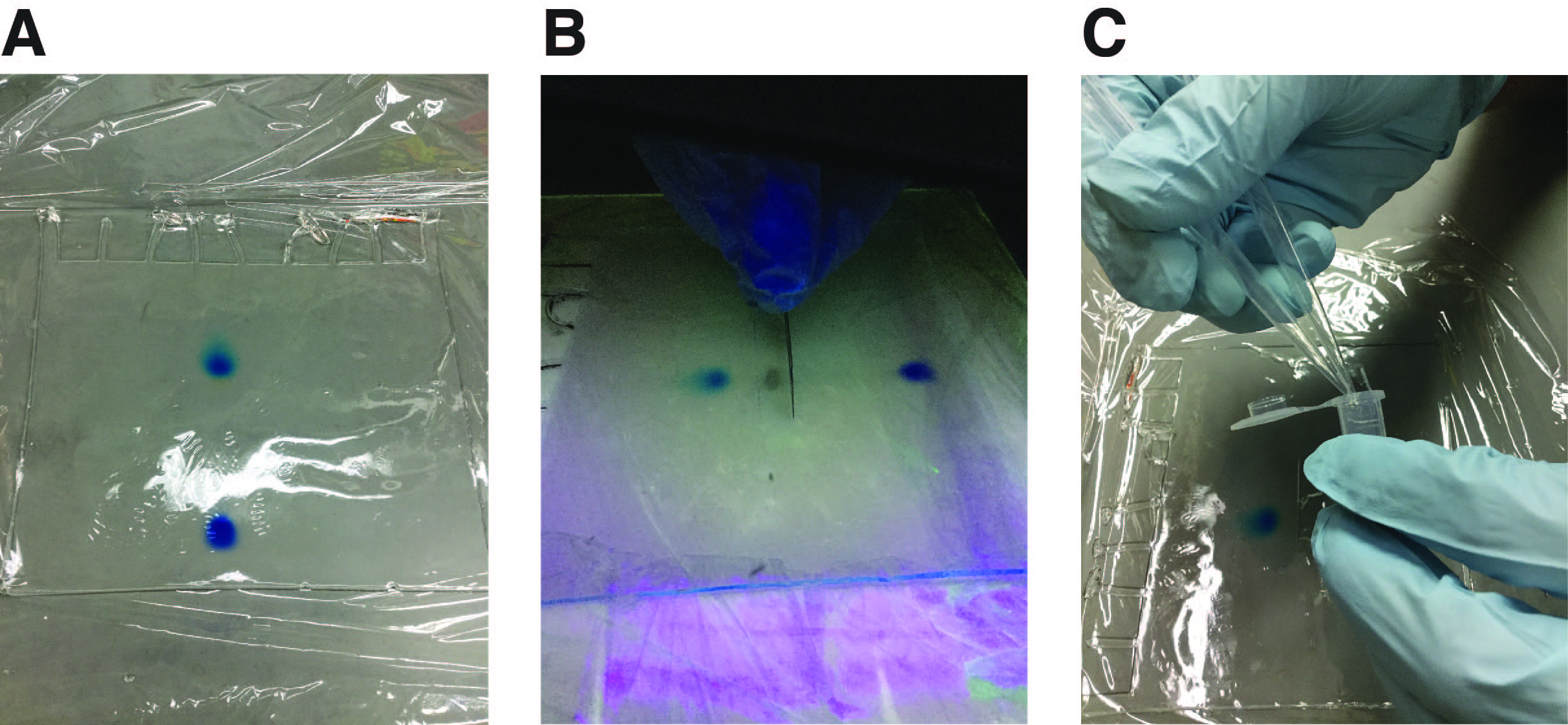
Figure 2. Workflow for RNA purification. A. The gel is transferred onto a plastic wrap after denaturing PAGE. B. Gel excision under UV light; C. A gel slice containing the RNA of interest is transferred into a tube with sterilized pipette tips. - Add 700 μl of PK buffer (see Recipes) and incubate the tube at room temperature (~25 °C) for 24 h on a rotator.
- Transfer the supernatant into a new 1.5 ml tube and add 700 μl of 2-propanol and mix by vortexing.
- Centrifuge at 17,000 x g at 4 °C for 20 min and remove the supernatant carefully.
- Add 700 μl of 70% ethanol.
- Centrifuge at 17,000 x g at 4 °C for 3 min and remove the supernatant.
- Repeat Steps B17 and B18.
- Air dry the pellet for 10 min and dissolve it in distilled water.
- Quantify the concentration of the RNA with a spectrophotometer.
- Transcribe reporter RNAs from DNA templates at 37 °C for 2 h using T7-Scribe Standard RNA IVT Kit following manufacturer’s instructions.
- RNA-dependent RNA polymerase (RdRP) activity assay
The flow chart of this assay is shown in Figure 3. After the reaction, the samples are electrophoresed on a urea gel or native gel. In urea PAGE, dsRNAs are denatured into two ssRNAs and migrate based on their molecular weights. Although RNase I treatment is essential for discrimination between RdRP products and TNTase products, this method allows the accurate estimation of the size of newly synthesized complementary strand. In contrast, because dsRNAs retain their double-helical structure in native PAGE, double-stranded RdRP products can be distinguished from single-stranded TNTase products based on their different mobilities without RNase I treatment. A drawback of native PAGE is that the mobility does not reflect the actual size of RNAs. The simultaneous use of both PAGE methods provides more accurate information for the enzymatic properties of RdRPs.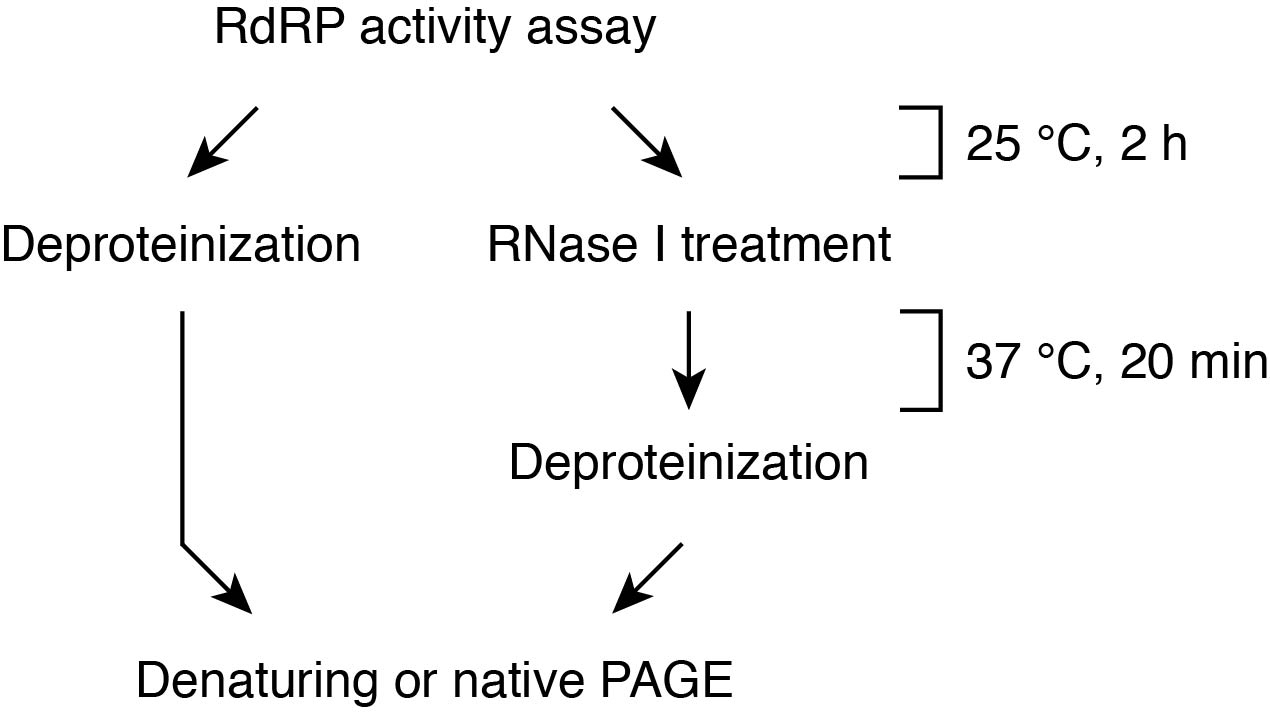
Figure 3. Flow chart of the procedure for in vitro RdRP assay- Combine the following reagents on ice in the order given: 20 μl of distilled water, 4 μl of 10x RdRP buffer (see Recipes), 4 μl of 5x diluted RNasin, 4 μl of 1 mM NTP mixture, 2 μl of 1 μM template RNA, 2 μl of α-32P-UTP (~3,000 Ci/mmol) and 4 μl of 100-200 nM RDR6.
- Incubate the tube at 25 °C for 2 h.
- Split the mixture into 4 tubes (tube A, B, A’ and B’) containing 10 μl each. Place them on ice.
- Add 111 μl of PK mixture (see Recipes) and 111 μl of PK mixture (Mg2+) (see Recipes) into tube A and tube A’, respectively (see Note 8). Vortex, and incubate at 25 °C until Step C6 is completed.
- Add 10 μl of RNase I mixture (see Recipes) into tube B and tube B’ (see Note 9) and incubate at 37 °C for 20 min.
- Add 111 μl of PK mixture and 111 μl of PK mixture (Mg2+) into tube B and tube B’, respectively, and vortex.
- Incubate all tubes at 65 °C for 20 min (see Note 10).
- Add 350 μl of 100% ethanol to each tube, vortex and centrifuge at 17,000 x g for 10 min at 4 °C. Remove the supernatant.
- Add 700 μl of 70% ethanol to each tube and centrifuge at 17,000 x g for 3 min at 4 °C. Remove the supernatant completely.
- Air dry the pellet for 10 min.
- For urea PAGE, dissolve the pellet (tube A and B) in 10 μl of 2x formamide dye and incubate at 95 °C for 5 min. For native PAGE, dissolve the pellet (tube A’ and B’) in 10 μl of 1x loading buffer (see Recipes) and keep the samples on ice.
- Prepare a urea gel containing 0.5x TBE and appropriate concentration of polyacrylamide or a native gel containing 0.5x TBE, 2 mM MgCl2 and appropriate concentration of polyacrylamide (see Note 11).
- Perform electrophoresis (see Notes 12, 13 and 14).
- Dry the gel using gel dryer under vacuum at 80 °C for 2 h.
- Expose the dried gel onto an imaging plate for ~12 h. Scan the plate using a laser scanner.
- Combine the following reagents on ice in the order given: 20 μl of distilled water, 4 μl of 10x RdRP buffer (see Recipes), 4 μl of 5x diluted RNasin, 4 μl of 1 mM NTP mixture, 2 μl of 1 μM template RNA, 2 μl of α-32P-UTP (~3,000 Ci/mmol) and 4 μl of 100-200 nM RDR6.
Data analysis
Here, we explain typical results of in vitro RdRP assay with the 100-nt RNA without the poly(A) tail. When total RNAs after the RdRP reaction are electrophoresed in a urea gel, multiple smeared bands are generally observed (Figure 4A RNase I-). At this time, these bands cannot be assigned to either TNTase or RdRP products. After ssRNA-specific RNase I treatment, some bands disappear and only a sharp band remains (Figure 4A RNase I+). This RNase I-resistant band is the RdRP product. In contrast, all the other bands erased by the RNase I treatment are single-stranded TNTase products (Figure 4A). When total RNAs after the RdRP reaction are electrophoresed in a native gel, double-stranded RdRP products migrate more slowly than single-stranded TNTase products (Figure 4B RNase I-), which can be validated by digestion of ssRNAs by RNase I treatment (Figure 4B RNase I+). Note that although the optimal concentration of RNase I is determined, RNase I treatment decreases dsRNA signals to some extent (Figure 4B). The signal of dsRNA products in the absence of RNase I treatment in native PAGE reflects the actual RdRP activity of RDR6.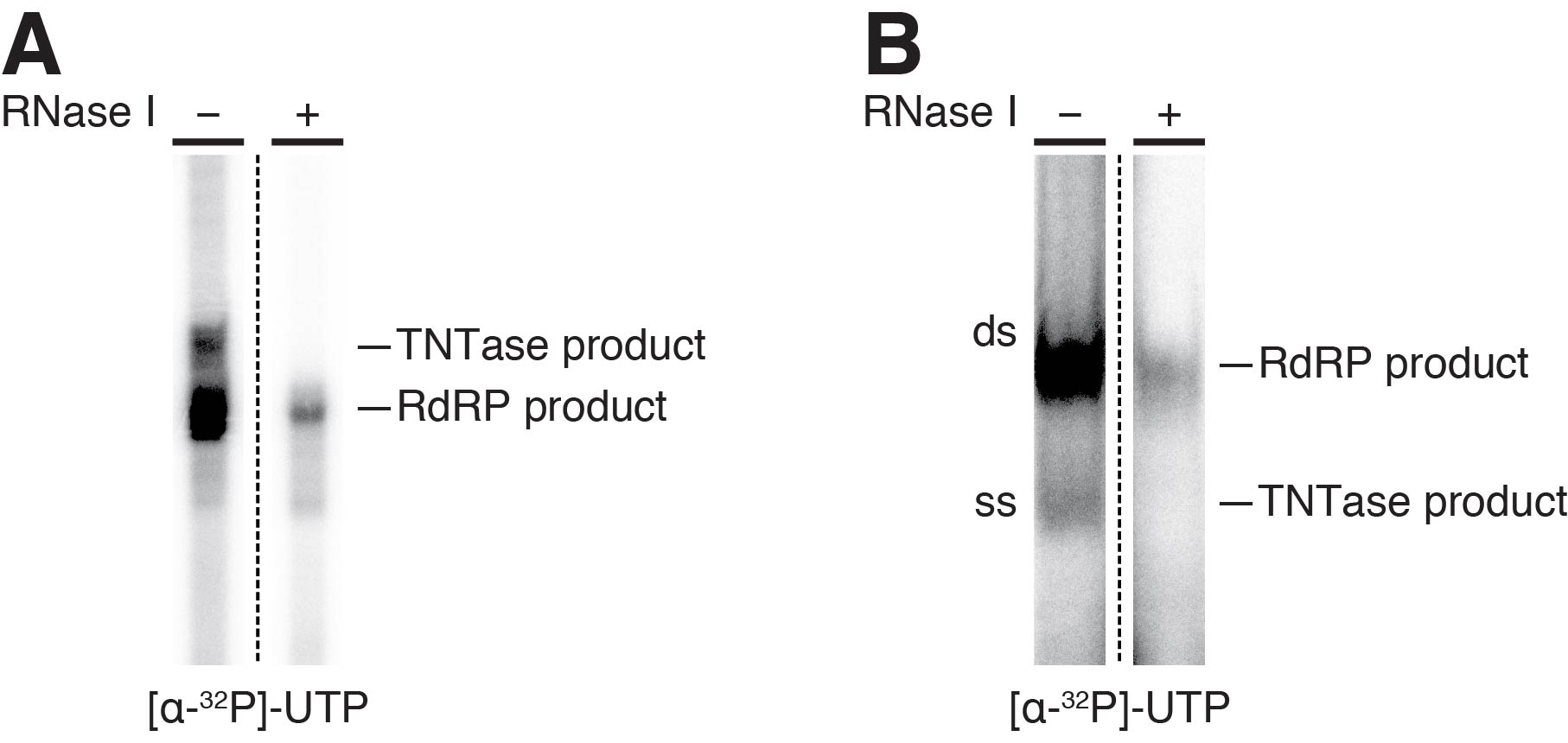
Figure 4. Representative results of in vitro RdRP assay. A. A typical banding pattern of RdRP- and TNTase-products from the 100-nt template RNA without the poly(A) tail on an 8% acrylamide-urea gel. B. A typical banding pattern of RdRP- and TNTase-products from the 100-nt template RNA without the poly(A) tail on a 4% acrylamide-native gel.
Notes
- S2 cells are statically cultured in a 100-mm dish with 10 ml of Schneider’s Drosophila medium containing 10% FBS and 1x antibiotic-antimycotic in an incubator at 27 °C. For maintenance of cells, passage S2 cells when the cell density reaches between 1.0 x 107 and 1.5 x 107 per ml, and split at a 1:10 dilution into a new 100-mm dish.
- To obtain high a concentration of recombinant RDR6, we usually prepare 500 ml of S2 cells (1.5 x 106 cells per ml). We aliquot it into 50 dishes (10 ml/dish) for transfection of pAFW-SUMO-AtRDR6. After expression of RDR6, S2 cells are collected into twelve 50-ml tubes (~40 ml each). After centrifugation, the cell pellet in each tube is resuspended with 300 μl of PBS, and transferred into a 1.5 μl tube. At Step A10, the washed magnetic beads in the 12 tubes are collected in a new tube. Then, 100 μl of 1x lysis buffer containing 1 mM DTT, 20% glycerol and 0.05 U per μl SUMOstar protease are added for elution of RDR6.
- To prepare the anti-FLAG antibody conjugated Dynabeads for immunopurification from e.g., 100 μl of S2 cell extract, 2 μl of anti-FLAG antibody and 100 μl slurry of Dynabeads are mixed in a tube by tapping, and incubated on a rotator at 4 °C for 1 h. Then, the tube is placed on a magnetic stand, and the supernatant is removed. The beads are washed once with 200 μl of 1x lysis buffer, and resuspended in 200 μl of 1x lysis buffer. The supernatant is removed just before incubation with S2 cell extract.
- We use SequaGel-UreaGel Concentrate to prepare urea gel.
- We usually use 5%, 8% and 15% acrylamide-urea gels for migration of 400-700 nt, 80-200 nt and < 50 nt RNAs, respectively.
- We resolve the RNAs at a constant voltage of 500 V in 0.5x TBE as running buffer.
- If the template RNAs are undetectable by UV shadowing, stain the gel with a nucleic acid staining dye for gel excision.
- For native PAGE samples (tube A’ and B’), we use ‘PK buffer (Mg2+)’ which contains magnesium ions but not EDTA to stabilize double-stranded RNAs (see Recipes).
- Although RNase I treatment allows discrimination of double-stranded RdRP products from single-stranded TNTase products, the use of a too high concentration of RNase I leads to degradation of dsRNAs. Therefore, the optimal concentration of RNase I should be carefully determined prior to the RdRP activity assay by using 32P body-labeled ss and dsRNA markers as substrates for RNase I.
- To avoid unwinding of dsRNAs, if the lengths of template RNAs are < 25 nt, it is better to incubate the RNA samples for native PAGE at 25 °C for 60 min or at 37 °C for 30 min in deproteinization step.
- We use 40(w/v)%-Acrylamide/Bis Mixed Solution(19:1) to prepare native gel.
- The running time should be optimized. The RdRP- and TNTase-products from the 100-nt template RNAs are well separated on an 8% acrylamide-urea gel (100 mm (H) x 150 mm (W) x 1 mm (D)) at a constant voltage of 500 V for 40 min or 4% acrylamide-native gel (100 mm (H) x 150 mm (W) x 1 mm (D)) at a constant voltage of 500 V for 40 min.
- To avoid spontaneous unwinding of double-stranded RNAs, keep the running buffer cold using plastic bars filled with coolant and perform electrophoresis in a cold room (Figure 5).
- To determine the relative size of the products, 5’ radiolabeled template RNAs can be used as the size markers.
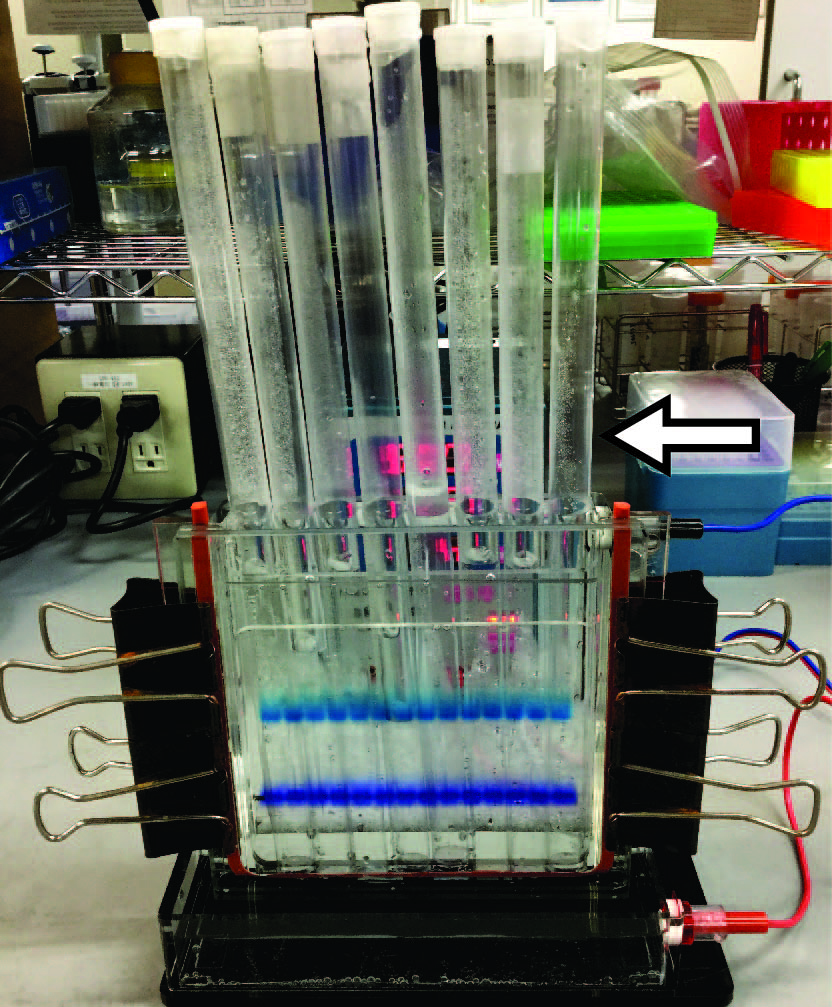
Figure 5. Representative image of native PAGE. The white arrow indicates a plastic bar filled with coolant.
Recipes
- Phosphate-buffered saline (PBS), pH 7.4
137 mM NaCl
2.7 mM KCl
10 mM Na2HPO4
1.8 mM KH2PO4 - Hypotonic lysis buffer
10 mM HEPES-KOH, pH 7.4
10 mM KCl
1.5 mM Mg(OAc)2
5 mM DTT
1x EDTA-free protease inhibitor cocktail - 1x lysis buffer
30 mM HEPES-KOH, pH 7.4
100 mM KOAc
2 mM Mg(OAc)2 - 2x formamide dye
10 mM EDTA, pH 8.0
98% (w/v) deionized formamide
0.025% (w/v) xylene cyanol
0.025% (w/v) bromophenol blue - PK buffer
100 mM Tris-HCl, pH 7.5
200 mM NaCl
2 mM EDTA, pH 8.0
1% (w/v) SDS - 1 mM NTP mixture
1 μl of 100 mM ATP in Ribonucleotide Solution set
1 μl of 100 mM UTP in Ribonucleotide Solution set
1 μl of 100 mM GTP in Ribonucleotide Solution set
1 μl of 100 mM CTP in Ribonucleotide Solution set
96 μl of distilled water - PK buffer (Mg2+)
100 mM Tris-HCl, pH 7.5
200 mM NaCl
1 mM MgCl2
1% (w/v) SDS - PK mixture
2 μl of glycogen
20 μl of Proteinase K
200 μl of PK buffer - PK mixture (Mg2+)
2 μl of glycogen
20 μl of Proteinase K
200 μl of PK buffer (Mg2+) - 5x TBE
0.446 M Tris
0.445 M H3BO3
0.01 M EDTA, pH 8.0 - 10x RdRP buffer
500 mM HEPES-KOH, pH 7.6
200 mM NH4OAc
80 mM MgCl2
1 mM EDTA, pH 8.0
20% (w/v) PEG 4000 - 2x RNase I buffer
100 mM Tris-HCl pH 8.0
600 mM NaCl
30 mM MgCl2 - RNase mixture
10 μl of 2x RNase I buffer
1 μl of RNase I
9 μl of distilled water - 2x loading buffer
4 mM MgCl2
0.5x TBE
50% (w/v) glycerol
0.04% (w/v) tartrazine
0.02% (w/v) bromophenol blue
0.02% (w/v) xylene cyanol
Acknowledgments
This protocol has been adapted and modified from Baeg et al., 2017 and Makeyev and Bamford, 2002. We are grateful to members of Tomari laboratory for comments on the manuscript. This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas (‘Nascent-chain Biology’) 26116003 (to H.I.), (‘Non-coding RNA neo-taxonomy’) 26113007 (to Y.T.), Grant-in-Aid for Young Scientists (A) 16H06159 (to H.I.), Grant-in-Aid for Challenging Exploratory Research 15K14444 (to H.I.) and Grant-in-Aid for JSPS Fellows 16J07290 (to K.B.). We declare no conflicting or competing interests.
References
- Aalto, A. P., Poranen, M. M., Grimes, J. M., Stuart, D. I. and Bamford, D. H. (2010). In vitro activities of the multifunctional RNA silencing polymerase QDE-1 of Neurospora crassa. J Biol Chem 285(38): 29367-29374.
- Baeg, K., Iwakawa, H. O. and Tomari, Y. (2017). The poly(A) tail blocks RDR6 from converting self mRNAs into substrates for gene silencing. Nat Plants 3: 17036.
- Curaba, J. and Chen, X. (2008). Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J Biol Chem 283(6): 3059-3066.
- Devert, A., Fabre, N., Floris, M., Canard, B., Robaglia, C. and Crete, P. (2015). Primer-dependent and primer-independent initiation of double stranded RNA synthesis by purified Arabidopsis RNA-dependent RNA polymerases RDR2 and RDR6. PLoS One 10(3): e0120100.
- Garcia-Ruiz, H., Takeda, A., Chapman, E. J., Sullivan, C. M., Fahlgren, N., Brempelis, K. J. and Carrington, J. C. (2010). Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22(2): 481-496.
- Li, H., Xu, L., Wang, H., Yuan, Z., Cao, X., Yang, Z., Zhang, D., Xu, Y. and Huang, H. (2005). The Putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell 17(8): 2157-2171.
- Makeyev, E. V. and Bamford, D. H. (2002). Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol Cell 10(6): 1417-1427.
- Mourrain, P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., Jouette, D., Lacombe, A. M., Nikic, S., Picault, N., Remoue, K., Sanial, M., Vo, T. A. and Vaucheret, H. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101(5): 533-542.
- Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L. and Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18(19): 2368-2379.
- Schneider, I. (1972). Cell lines derived from late embryonic stages of Drosophila melanogaster. J Embryol Exp Morphol 27(2): 353-365
- Volpe, T. A., Kidner, C., Hall, I. M., Teng, G., Grewal, S. I. and Martienssen, R. A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297(5588): 1833-1837.
- Wang, X. B., Wu, Q., Ito, T., Cillo, F., Li, W. X., Chen, X., Yu, J. L. and Ding, S. W. (2010). RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc Natl Acad Sci U S A 107(1): 484-489.
- Xie, Z., Johansen, L. K., Gustafson, A. M., Kasschau, K. D., Lellis, A. D., Zilberman, D., Jacobsen, S. E. and Carrington, J. C. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2(5): E104.
- Yu, D., Fan, B., MacFarlane, S. A. and Chen, Z. (2003). Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol Plant Microbe Interact 16(3): 206-216.
- Zong, J., Yao, X., Yin, J., Zhang, D. and Ma, H. (2009). Evolution of the RNA-dependent RNA polymerase (RdRP) genes: duplications and possible losses before and after the divergence of major eukaryotic groups. Gene 447(1): 29-39.
Article Information
Copyright
© 2018 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Baeg, K., Tomari, Y. and Iwakawa, H. (2018). In vitro RNA-dependent RNA Polymerase Assay Using Arabidopsis RDR6. Bio-protocol 8(1): e2673. DOI: 10.21769/BioProtoc.2673.
Category
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Isolation and purification
Plant Science > Plant molecular biology > RNA
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









