- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparation of Onion Epidermal Cell Walls for Imaging by Atomic Force Microscopy (AFM)
Published: Vol 7, Iss 24, Dec 20, 2017 DOI: 10.21769/BioProtoc.2647 Views: 15102
Reviewed by: Xinyan ZhangKirsten KnoxAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2312 Views

Live-Cell Monitoring of Piecemeal Chloroplast Autophagy
Masanori Izumi [...] Shinya Hagihara
Nov 5, 2025 1688 Views

Chloroplast Movement Imaging Under Different Light Regimes With a Hyperspectral Camera
Paweł Hermanowicz [...] Justyna Łabuz
Dec 20, 2025 749 Views
Abstract
The growing plant cell wall is comprised of long, thin cellulose microfibrils embedded in a hydrated matrix of polysaccharides and glycoproteins. These components are typically constructed in layers (lamellae) on the inner surface of the cell wall, i.e., between the existing wall and the plasma membrane. The organization of these components is an important feature for plant cell growth and mechanics. To directly visualize the nano-scale structure of the newly-deposited surface of primary plant cell walls without dehydration or chemical extraction, a protocol of cell wall preparation for AFM imaging the most recently-synthesized cell wall surface in aqueous solutions was developed. Although the method was developed for onion scale epidermal peels, it can also be adapted to other organs, such as Arabidopsis hypocotyls, as well as ground samples of cell walls from the leaf petioles or hypocotyls of Arabidopsis and cucumber, maize coleoptiles and onion parenchyma. Potential artifacts of AFM imaging of plant cell walls are also discussed.
Keywords: Atomic force microscopyBackground
The structure of primary plant cell walls plays a key role in determining cell wall biomechanical properties and regulating plant cell growth and morphogenesis (Cosgrove, 2005 and 2016). To visualize cell wall organization, common methods include transmission and scanning electron microscopy (TEM, SEM), light microscopy and AFM (McCann et al., 1990; Marga et al., 2005; Anderson et al., 2010; Ding et al., 2012; Abraham and Elbaum, 2013; Zhang et al., 2014; Zheng et al., 2017). In recent years AFM has enabled the imaging of soft biological samples in fluid, thus allowing studies of plant cell walls at nm-resolution in a close-to-native state, without dehydration, harsh chemical treatment, embedding or sectioning. Compared to EM techniques, which typically require dehydrated samples for high resolution imaging, AFM avoids dehydration artifacts and simplifies the sample preparation procedure while achieving high resolution imaging at the nanometer scale (Zhang et al., 2016). Combining AFM imaging with enzyme treatments, mechanical testing or use of cell wall mutants (Xiao et al., 2016; Zhang et al., 2017), we can test the hypothesized roles of specific cell wall components in cell growth, mechanics and cell wall structure.
For this protocol we focus on the onion scale epidermal cell wall, which has been the subject of numerous mechanical, spectroscopic and microscopic studies (Wilson et al., 2000; Kerstens et al., 2001; Hepworth and Bruce, 2004; Vanstreels et al., 2005; Loodts et al., 2006; Suslov et al., 2009). A key difference between our preparation method and that of previous authors is that our method splits open the epidermal cell layer, separating the outer epidermal cell wall from the remainder of the epidermal cell. After a brief wash to remove membranes and cellular debris, the newly-synthesized surface is ready for direct imaging by AFM. This is possible because the abaxial epidermis (on the convex or outer surface of the onion scale) adheres tightly to the underlying parenchyma tissues, so the peeling procedure splits open the epidermal cells. In previous studies with onion, whole-cell epidermal layers were peeled from the adaxial (inner or concave) surface of the onion scale, i.e., this cell layer adheres weakly to the underlying tissues and so separates intact at the interface (the middle lamella) with the underlying tissue. This difference in peeling procedure is of course important for surface-imaging methods such as AFM and SEM.
Materials and Reagents
- Razor blades
- Double sided tape (Permanent Double Sided Tape) (Scotch, catalog number: 3136 )
- Glass microscope slide (75 x 25 mm) (VWR, catalog number: 48300-025 )
- Charged microscope slide (75 x 25 mm) (VWR, catalog number: 48312-905 )
- 0.2 µm filter (Whatman GD/X 25 mm Sterile PVDF Syringe Filter) (GE Healthcare, catalog number: 6900-2502 )
- Fresh onions bulbs (Allium cepa, cv. Cometa), approximately 8 cm in diameter
- Nail polish (nitrocellulose dissolved in butyl and ethyl acetate)
- 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid sodium salt (HEPES) (Sigma-Aldrich, catalog number: H7006 )
- Tween-20 (Sigma-Aldrich, catalog number: P9416 )
- Sodium acetate, anhydrous (Sigma-Aldrich, catalog number: S2889 )
- Washing buffer (see Recipes)
- Imaging buffer (see Recipes)
Equipment
- Forceps
- Bruker Dimension Icon atomic force microscope with ScanAsyst and PeakForce QNM (Quantitative Nanomechanical Property Mapping) operation package
- Scanasyst-fluid + AFM tips (Bruker, CA)
- Rocking platform (VWR, model: Model 200 )
- Slide warmer (50 °C) (Fisher Scientific, model: Model 77 , catalog number: 12-594)
Software
- Nanoscope for AFM operation and Nanoscope Analysis for image analysis (Bruker)
Procedure
- Purchase fresh onion bulbs (Allium cepa, cv. Cometa or similar white varieties, ~8 cm in diameter) from a local grocery store. The onions can be stored at 4 °C up to two weeks.
- Remove the dry, papery outer layers. The fleshy scales are numbered consecutively 1, 2, 3…, n with scale #1 being the oldest, outermost layer (Figure 1A).
- Excise scale #5 as a ~1 cm wide strip and press a 2 x 1.2 cm piece of double-sided tape to the convex, abaxial surface of the upper middle (near the tip) part of the onion scale (Figure 1B).
- Peel off the epidermis (with the tape) by hand, trim to ~1.5 x 1.2 cm (Figure 1C), and place the tape and the sample onto a clean glass slide, with the sample side facing up. Press the perimeter of the sample onto the slide.
- Seal the perimeter of the sample with a narrow line of nail polish and add a droplet (~20 μl) of washing buffer (see Recipes) to the center of the sample to keep it from drying (Figure 1D). The washing buffer should not submerge the surrounding still-wet nail polish. Steps 2 to 5 should be completed in ~30 sec.

Figure 1. Procedure of onion epidermal wall preparation for AFM. A. Numbered onion scales with scale #1 being the oldest, outermost layer. Epidermal peels from scale #5 were usually used for imaging, although other scales are compatible with this procedure. B. A ~1 cm wide strip was excised and a double-sided tape was pressed against the abaxial side of the upper middle (near the tip) part of the onion scale. C. The epidermis was peeled off along with the tape. D. The epidermis and the tape were pressed onto the slide and a drop of washing buffer was added to the center of the sample.
- Allow the nail polish to cure for ~2 min or longer before adding more of the same washing buffer (~100 μl) and gently agitate on a rocking platform for at least 20 min. The slide is put in a Petri dish with lid closed to reduce evaporation. Before scanning, examine the peel under an optical microscope to verify that epidermal wall indeed separated from the rest of the cell, exposing the inner surface of the wall for direct contact with the AFM tip.
Note: In our experience, peeling of the abaxial epidermis results in separation of the outer (periclinal) cell wall from the rest of the epidermal cell layer. However, this is not always uniform and must be verified by examination of the peel by optical microscopy after the detergent washing step. Cells with serrated edge or teeth-like features are usually torn open and good candidates for AFM imaging (Figure 2). The nucleus and other organelles should be absent. For AFM observations it is not essential that all of the cells split open in this way. Regions of the peel with intact cell walls can be ignored for the AFM imaging. The advantage of the onion scale abaxial epidermis is that it generally peels in this manner quite reproducibly and uniformly. Epidermal layers from other tissues sometimes split open in a similar fashion, others not. This must be determined by trial and error.

Figure 2. Optical micrograph showing an AFM tip hovering above an epidermal cell. The serrated, teeth-like structures are good indications that the epidermal cells are split open.
- Calibrate the deflection sensitivity (~50-60 nm/V) and spring constant (0.2-0.7 N/m) of the AFM tip with a clean glass slide in fluid, following the manufacturer’s protocol. The tip radius (~1.8 nm), tip half angle (17.5°), and sample Poisson’s ratio (estimated as 0.3) need to be entered manually. The values given here are based on the tips that we routinely use (Scanasyst-fluid+ AFM tips) and may differ for other tips.
- Throughout the scanning processes, the samples are immersed in imaging buffer (20 mM HEPES buffer [pH 7.0] or 20 mM sodium acetate [pH 4.5], see Recipes).
- Start scanning at 2 x 2 μm size and 512 x 512 sampling rates (thus resolution of ~4 nm/pixel). For high resolution images, scan the sample at 0.5 x 0.5 μm or smaller. The scanning parameters are not identical for all materials, but typically fall in the following ranges, which are determined empirically: a peak force set point (600 pN-1.5 nN), scan rate (0.4-0.7 Hz), peak force frequency (1 kHz), tip velocity (0.9-1.5 µm/sec). The gain is auto controlled by the software but the set point and scan rate are typically controlled by the user for consistency throughout imaging (Figure 3).
- For standard cell wall imaging, scan at least five different cells from each sample and choose representative images for further analysis.
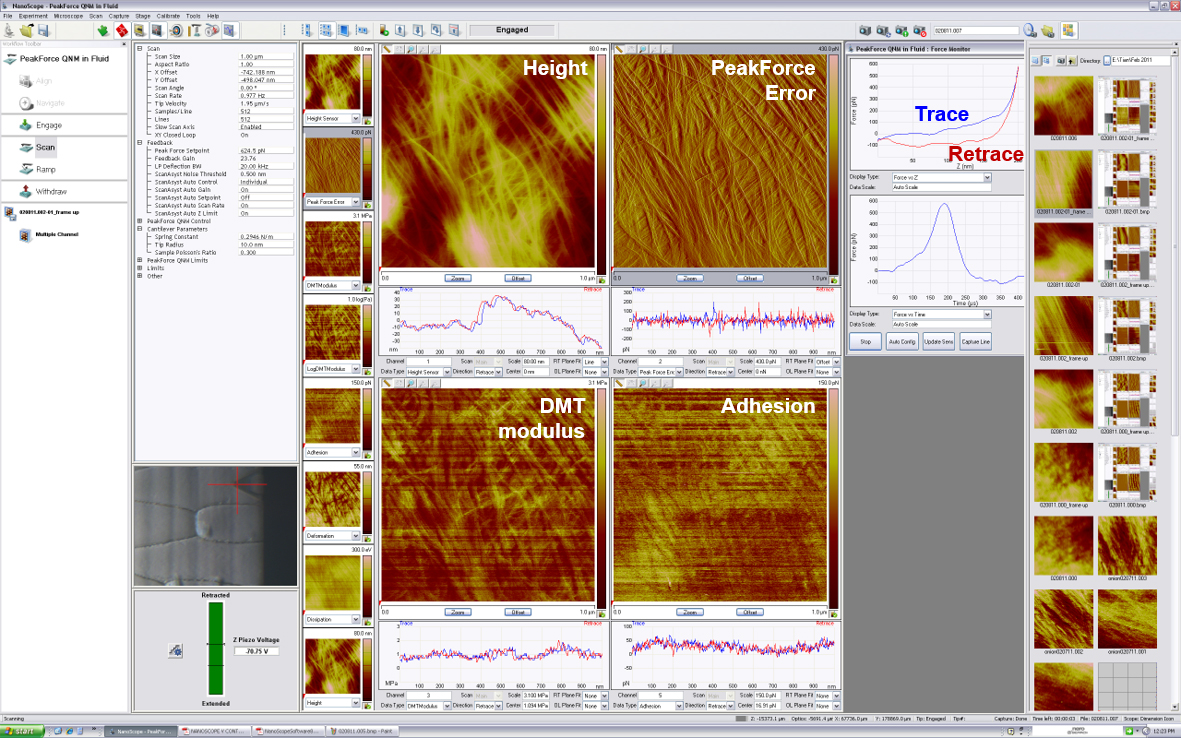
Figure 3. A screenshot of Nanoscope interface during AFM operation. Four different channels are in display: Height, PeakForce Error, DMT modulus and Adhesion. Trace-retrace curves of each scan lines are displayed under each channel. The real-time force-distance curve is shown on the upper right corner.
Data analysis
In Nanoscope Analysis software, the images are usually flattened to remove tilt or bow in each scan line and are exported in TIFF format. To export gray scale images, choose color table 0. For representative data see Zhang et al. (2014).
Notes
- All buffers must be of highest purity, using ddH2O and filtering through a 0.2 µm filter.
- Although much of our work is with onion scale #5, walls from other scales may also be used and studied in parallel with optical techniques (Kafle et al., 2014).
- While acquiring images, the real-time force curve should be monitored to optimize contact of tip to surface, hence generating a better-resolved image and more accurate mechanical mapping data. Also, the trace-retrace curves being displayed with each channel should be watched for indications of movement of the sample or poor tip-surface interaction. If the sample is mechanically unstable or if the tip does not make good contact with the surface, the resolution may be poor or imaging artifacts could be generated.
- To prepare walls from onion parenchyma, peel off the abaxial and adaxial side of epidermal layers, and grind the parenchyma to fine powder in liquid nitrogen. Wash the cell wall pellet with 20 mM HEPES buffer (pH 7.0) and 0.1% Tween-20 three times or until the supernatant is clear (centrifuge at 1,500 x g for 3 min). Resuspend the wall fragments in 20 mM HEPES buffer and add a droplet of the sample to a clean charged glass slide. Evaporate excess water on a slide warmer (50 °C) until the sample is still visibly moist but adheres to the slide (~15 min). Rinse the slide immediately with 20 mM HEPES buffer to remove loosely bound fragments before AFM scanning. We assume the middle lamella remains intact (Zamil et al., 2014) and so the only exposed wall surfaces are the inner faces of the cell walls. This assumption may not be true for other tissues.
- A similar grinding procedure can be used for Arabidopsis thaliana (Columbia) petioles (4-week-old; day/night, 16 h/8 h; temperature, 22 °C/16 °C), Arabidopsis dark grown hypocotyls (3 day at 22 °C), cucumber (Cucumis sativus) hypocotyls and maize (Zea mays) coleoptiles (dark grown for 4 days at 27 °C). Epidermal cell walls are identified by their characteristic appearance under the light microscope (elongated, rectangular shape, Figure 4). The cuticle side can be identified from the smoothness of the acquired image and the lack of fibrillar structures.
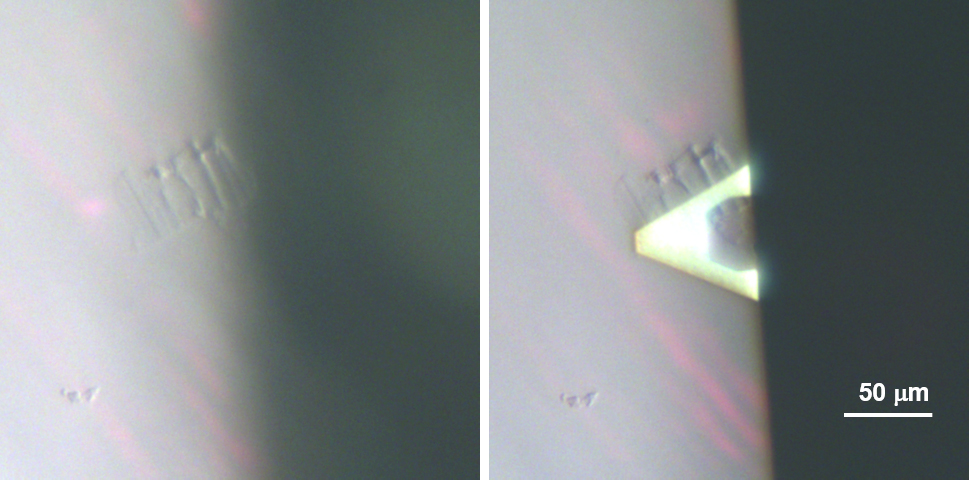
Figure 4. A light micrograph showing the rectangular shaped epidermal fragment for AFM scanning
- Beware of possible artifacts induced by a dulled AFM tip or drifting due to temperature fluctuations or sample movement. The AFM tip can be contaminated or damaged during manufacturing process or during scanning, resulting in images with less sharp or ghosting features (Figure 5). Drifting is a common problem and should be examined in the beginning of the experiment by disabling ‘the slow axis scan’ (an option in scanning parameters) and repeatedly scanning one line. If drifting exists, the imaging features will appear tilted rather than vertically straight (Figure 6). Drifting can be limited by stabilizing samples in fluid for 1 or 2 h before collecting images. If the problem persists, scan a different cell or another sample.
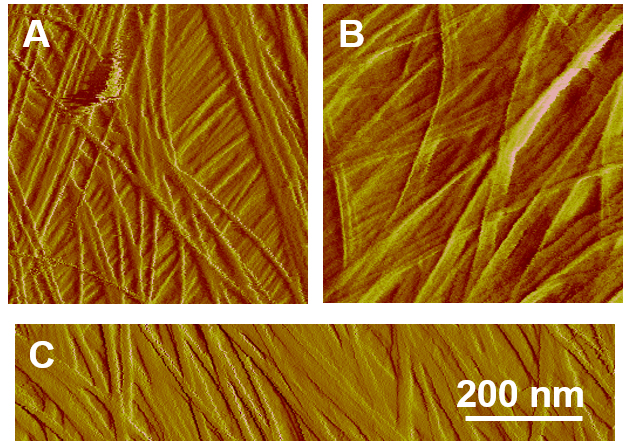
Figure 5. Common artifacts of AFM images due to dulled or broken tip. A. A representative AFM image of onion epidermis under normal conditions; B. Doubling/ghosting features of an AFM image due to a broken tip; C. Artefactual widening of microfibrils caused by a dulled tip.
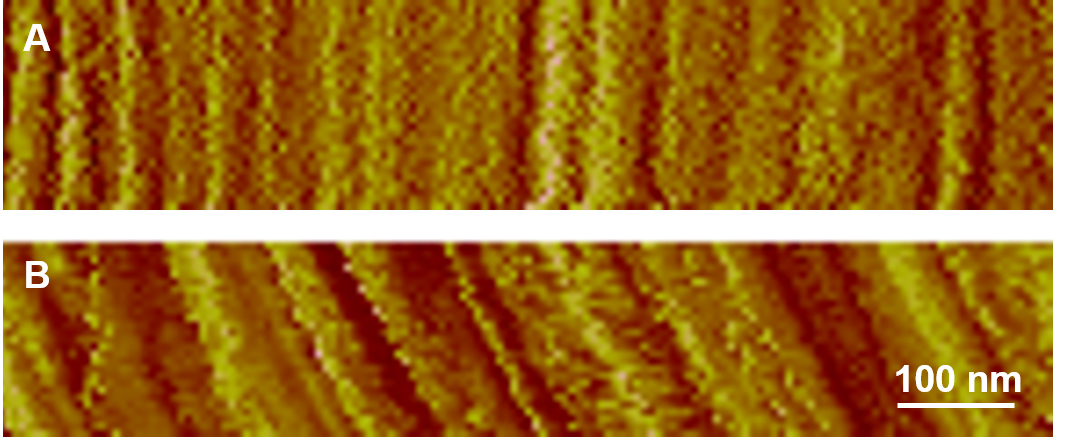
Figure 6. Disabling slow axis scan to check for drifting. A. Vertically straight features indicate there is no or very little drift. B. Tilted lines indicate drift.
Recipes
- Washing buffer
20 mM HEPES with 0.1% Tween-20, pH 7.0
- Imaging buffer
20 mM HEPES (pH 7.0) or 20 mM sodium acetate (pH 4.5)
Acknowledgments
This protocol was adapted from previous work (Zhang et al., 2014; Zhang et al., 2016). We thank Ed Wagner and Liza Wilson for technical assistance and AFM maintenance and Dr. Sarah Kiemle for comments on the manuscript. This work was supported by the Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences (grant No. DE-SC0001090). The authors declare no conflict of interest.
References
- Abraham, Y. and Elbaum, R. (2013). Quantification of microfibril angle in secondary cell walls at subcellular resolution by means of polarized light microscopy. New Phytol 197(3): 1012-1019.
- Anderson, C. T., Carroll, A., Akhmetova, L. and Somerville, C. (2010). Real-time imaging of cellulose reorientation during cell wall expansion in Arabidopsis roots. Plant Physiol 152(2): 787-796.
- Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat Rev Mol Cell Biol 6(11): 850-861.
- Cosgrove, D. J. (2016). Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J Exp Bot 67(2): 463-476.
- Ding, S. Y., Liu, Y. S., Zeng, Y., Himmel, M. E., Baker, J. O. and Bayer, E. A. (2012). How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science 338: 1055-1060.
- Hepworth, D. G and Bruce, D. M. (2004). Relationships between primary plant cell wall architecture and mechanical properties for onion bulb scale epidermal cells. J Texture Stud 35: 586-602.
- Kafle, K., Xi, X. N., Lee, C. M., Tittmann, B. R., Cosgrove, D. J., Park, Y. B. and Kim, S. H. (2014). Cellulose microfibril orientation in onion (Allium cepa L.) epidermis studied by atomic force microscopy (AFM) and vibrational sum frequency generation (SFG) spectroscopy. Cellulose 21: 1075-1086.
- Kerstens, S., Decraemer, W. F. and Verbelen, J. P. (2001). Cell walls at the plant surface behave mechanically like fiber-reinforced composite materials. Plant Physiol 127(2): 381-385.
- Loodts, J., Tijskens, E., Wei, C. F., Vanstreels, E., Nicolai, B. and Ramon, H. (2006). Micromechanics: Simulating the elastic behavior of onion epidermis tissue. J Texture Stud 37: 16-34.
- Marga, F., Grandbois, M., Cosgrove, D. J. and Baskin, T. I. (2005). Cell wall extension results in the coordinate separation of parallel microfibrils: evidence from scanning electron microscopy and atomic force microscopy. Plant J 43(2): 181-190.
- McCann, M. C., Wells, B. and Roberts, K. (1990). Direct visualization of cross-links in the primary plant cell wall. J Cell Sci 96: 323-334.
- Suslov, D., Verbelen, J. P. and Vissenberg, K. (2009). Onion epidermis as a new model to study the control of growth anisotropy in higher plants. J Exp Bot 60(14): 4175-4187.
- Vanstreels, E., Alamar, A. C., Verlinden, B. E., Enninghorst, A., Loodts, J. K. A., Tijskens, E., Ramon, H. and Nicolai, B. M. (2005). Micromechanical behaviour of onion epidermal tissue. Postharvest Biol Tec 37: 163-173.
- Wilson, R. H., Smith, A. C., Kacurakova, M., Saunders, P. K., Wellner, N. and Waldron, K. W. (2000). The mechanical properties and molecular dynamics of plant cell wall polysaccharides studied by Fourier-transform infrared spectroscopy. Plant Physiol 124(1): 397-405.
- Xiao, C., Zhang, T., Zheng, Y., Cosgrove, D. J. and Anderson, C. T. (2016). Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol 170(1): 234-249.
- Zamil, M. S., Yi, H. and Puri, V. M. (2014). Mechanical characterization of outer epidermal middle lamella of onion under tensile loading. Am J Bot 101: 778-787.
- Zhang, T., Mahgsoudy-Louyeh, S., Tittmann, B. and Cosgrove, D. J. (2014). Visualization of the nanoscale pattern of recently-deposited cellulose microfibrils and matrix materials in never-dried primary walls of the onion epidermis. Cellulose 21: 853-862.
- Zhang, T., Vavylonis, D., Durachko, D. M. and Cosgrove, D. J. (2017). Nanoscale movements of cellulose microfibrils in primary cell walls. Nat Plants 3: 17056.
- Zhang, T., Zheng, Y. and Cosgrove, D. J. (2016). Spatial organization of cellulose microfibrils and matrix polysaccharides in primary plant cell walls as imaged by multichannel atomic force microscopy. Plant J 85(2): 179-192.
- Zheng, Y., Cosgrove, D. J. and Ning, G. (2017). High-resolution field emission scanning electron microscopy (FESEM) imaging of cellulose microfibril organization in plant primary cell walls. Microsc Microanal 23(5): 1048-1054.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Zhang, T. and Cosgrove, D. J. (2017). Preparation of Onion Epidermal Cell Walls for Imaging by Atomic Force Microscopy (AFM). Bio-protocol 7(24): e2647. DOI: 10.21769/BioProtoc.2647.
Category
Plant Science > Plant cell biology > Cell imaging
Cell Biology > Cell imaging > Atomic force microscopy
Biochemistry > Carbohydrate > Cellulose
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










