- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Stationary-phase Mutagenesis Soft-agar Overlay Assays in Bacillus subtilis
Published: Vol 7, Iss 23, Dec 5, 2017 DOI: 10.21769/BioProtoc.2634 Views: 7416
Reviewed by: Valentine V TrotterAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Novel Method of Inducible Directed Evolution to Evolve Complex Phenotypes
Ibrahim S. Al’Abri [...] Nathan Crook
Oct 20, 2022 3062 Views

Comprehensive Mapping of EZ-Tn5 Transposon Insertion Sites in Pseudomonas argentinensis SA190 Using RATE-PCR
Büsra Elkatmis [...] Maged M. Saad
Jul 20, 2025 1751 Views

Editing the Serratia proteamaculans Genome Using the Allelic Exchange Method
Ksenia Chukhontseva [...] Ilya Demidyuk
Sep 20, 2025 1387 Views
Abstract
Elucidating how a population of non-growing bacteria generates mutations improves our understanding of phenomena like antibiotic resistance, bacterial pathogenesis, genetic diversity and evolution. To evaluate mutations that occur in nutritionally stressed non-growing bacteria, we have employed the strain B. subtilis YB955, which measures the reversions rates to the chromosomal auxotrophies hisC952, metB5 and leuC427 (Sung and Yasbin, 2002). This gain-of-function system has successfully allowed establishing the role played by repair systems and transcriptional factors in stress-associated mutagenesis (SPM) (Barajas-Ornelas et al., 2014; Gómez-Marroquín et al., 2016). In a recent study (Castro-Cerritos et al., 2017), it was found that Ribonucleotide Reductase (RNR) was necessary for SPM; this enzyme is essential in this bacterium. We engineered a conditional mutant of strain B. subtilis YB955 in which expression of the nrdEF operon was modulated by isopropyl-β-D-thiogalactopyranoside (IPTG) (Castro-Cerritos et al., 2017). The conditions to determine mutation frequencies conferring amino acid prototrophy in three genes (hisC952, metB5, leuC427) under nutritional stress in this conditional mutant are detailed here. This technique could be used to evaluate the participation of essential genes in the mutagenic processes occurring in stressed B. subtilis cells.
Keywords: Bacillus subtilisBackground
Around 270 genes, including dnaA, dnaB, dnaC, involved in DNA replication as well as the nrdEF operon, which encodes RNR, are considered essential for B. subtilis growth (Kobayashi et al., 2003). Here we describe a protocol that has been applied to understand the role of this enzyme in modulating events of mutagenesis in nutritionally stressed non-growing cells of strain B. subtilis YB955 (hisC952, lecuC427, metB5). To this end, we implemented a genetic system that represses the expression of the essential nrdEF operon (non-permissive condition) while limiting two of the essential amino acids to avoid transient and weak phenotypes produced by variants of less efficient enzymes (Pybus et al., 2010). These restriction conditions allowed us first inquiring whether RNR influences mutagenesis in a population of non-growing (but viable) B. subtilis cells. In a second stage, a soft agar overlay, which provides permissive conditions for growth, allows detection of prototrophic colonies only if a mutation that restores at least one amino acid prototrophy has occurred. Whereas this protocol has been employed here with RNR, it can be potentially extended to study additional genes that are essential for metabolism as well as DNA replication and transcription. In addition, conditions may be adjusted to other bacterial species or selective markers.
Materials and Reagents
- 15- and 50-ml sterile Falcon tubes. Conical Centrifuge Tubes (Corning, Axygen®, catalog numbers: SCT-15ML-500 ; SCT-50ML-500 )
- Pasteur pipettes
- Disposable (90 mm diameter) Petri dishes (BD, catalog number: 252777 )
- Cellulose filter paper 0.22 µm (Merck, catalog number: GSWP04700 )
- Bacillus subtilis strains YB955 (hisC952 metB5 leuC427 xin-1 Spβs) and PERM1017 (hisC952 metB5 leuC427 ΔnrdE::lacZ Pspac-nrdEF::ery Eryr)
- Penassay broth (PAB; antibiotic medium 3; Difco Laboratories, Sparks, MD) (Fisher Scientific, catalog number: DF0243-17-8)
Manufacturer: BD, catalog number: B224320 . - Isopropyl-β-D-thiogalactoside (IPTG) (Promega, catalog number: V3951 )
- L-Leucine (Sigma-Aldrich, catalog number: L8000 )
- L-Glutamic acid (Sigma-Aldrich, catalog number: 49449 )
- L-Isoleucine (Sigma-Aldrich, catalog number: 58879 )
- L-Methionine (Sigma-Aldrich, catalog number: M9625 )
- L-Histidine monohydrochloride monohydrate (Sigma-Aldrich, catalog number: 53370 )
- Ammonium sulfate ((NH4)2SO4) (Karal, catalog number: 3016 )
- Potassium phosphate dibasic (K2HPO4) (Karal, catalog number: 5080 )
- Potassium phosphate monobasic (KH2PO4) (Karal, catalog number: 5079 )
- Sodium citrate (C6H5Na3O7·2H2O) (Avantor Performance Materials, J.T. Baker®, catalog number: 3646-01 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Karal, catalog number: 6056 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Karal, catalog number: 6054 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C2536 )
Note: This product has been discontinued. - Ferrous chloride tetrahydrate (FeCl2·4H2O) (Avantor Performance Materials, J.T. Baker®, catalog number: 2064-01 )
- Zinc chloride (ZnCl2) (Sigma-Aldrich, catalog number: Z0152 )
- Copper(II) chloride dihydrate (CuCl2·2H2O) (Karal, catalog number: 8021 )
- Cobalt(II) chloride hexahydrate (CoCl2·6H2O) (Karal, catalog number: 8026 )
- Sodium molybdate dehydrate (Na2MoO4·2H2O) (Karal, catalog number: 4072 )
- Agar (BD, catalog number: 215000 )
- Dextrose (BD, catalog number: 216800 )
- PAB medium (PAB) (see Recipes)
- Amino acid solutions (see Recipes)
- 1 M IPTG (see Recipes)
- 10x Spizizen salts (10x SS)
- 100x trace elements (see Recipes)
- Spizizen minimal salts (SS) (see Recipes)
- Spizizen minimal medium (SMM) (see Recipes)
- Soft agar (see Recipes)
Equipment
- Orbital shaker Lab-Line MaxQ 4000 (Thermo Fisher Scientific, Thermo ScientificTM, model: MaxQTM 4000 )
- Conventional incubator set at 37 °C
- Spectrophotometer (Biochrom, model: WPA CO7500 )
- Centrifuge Thermo Scientific IEC CL30R (Thermo Fisher Scientific, Thermo ScientificTM, model: IEC CL30R )
- Conventional autoclave
- 125-ml sterile Erlenmeyer flasks
- Reusable syringe filter holders (Merck, catalog number: SX0002500 )
- Magnetic stirring bar
Procedure
- Cell propagation
- Propagate a 1:50 dilution of an overnight culture of B. subtilis strains in 25 ml of PAB medium (supplemented with 0.1 mM IPTG, if necessary, see Recipes). Incubate at 37 °C with vigorous shaking (250 rpm), monitor growth by taking periodical readings on a spectrophotometer set at 600 nm.
- Plot OD600 nm versus time in a semi-log sheet and identify time zero (T0), defined as the time point in the culture when the slopes of the logarithmic and stationary phases of growth intercept (see Supplemental Figure S1).
- 90 min after T0, transfer 10 ml of the culture to a sterile 15-ml Falcon tube and centrifuge (4, 800 x g; 10 min, 23 °C); wash cell pellets 3 times each with 10 ml of 1x SS (see Recipes) and resuspend in an equal volume of the same solution.
- Propagate a 1:50 dilution of an overnight culture of B. subtilis strains in 25 ml of PAB medium (supplemented with 0.1 mM IPTG, if necessary, see Recipes). Incubate at 37 °C with vigorous shaking (250 rpm), monitor growth by taking periodical readings on a spectrophotometer set at 600 nm.
- Viable counts
Determine the initial number of bacteria for each strain by preparing serial ten-fold dilutions in 1x SS, plating dilutions 10-5 to 10-7 on SMM supplemented with histidine, methionine and leucine, incubating for 48 h at 37 °C and scoring the number of CFU per ml. - Selective pressure
- Take aliquots of 1 ml of the cell suspension and, independently, transfer to three 50-ml Falcon tubes containing 49 ml of soft agar SA (one tube with SA1 and two for SA2) pre-warmed at 42 °C.
- Decant 4 ml of the final suspension onto each of 12 plates of SMM-His-Leu- and 24 plates of SMM-Met-Leu-, allow to solidify and incubate the plates at 37 °C for 10 days. Of note, supplement plates with 1 mM IPTG to activate nrdEF expression in strain B. subtilis PERM1017.
Note: The combination (His-, Met-) in addition to the absence of three amino acids (His-, Leu-, Met-) is not used to avoid favor suppressor mutations that have been suggested revert His and Met auxotrophies simultaneously (Sung and Yasbin, 2002).
- Take aliquots of 1 ml of the cell suspension and, independently, transfer to three 50-ml Falcon tubes containing 49 ml of soft agar SA (one tube with SA1 and two for SA2) pre-warmed at 42 °C.
- Colony growth
- During days 2, 4, 6, 8, and 10, pick two plates of His- Leu- medium and four plates of Met- Leu-, proceed to prepare soft agar to obtain His- (SA3), Met- (SA4) or Leu- (SA5) media (see Recipes). Over these SMM plates pour a first overlay with the cells and a second overlay with the selective medium (Figure 1 and Supplemental Figure S2). This results in single amino acid dropout media that selects for His+, Met+ or Leu+ revertants. Incubate the plates at 37 °C and record the number of revertants His+, Met+, and Leu+ colonies that arise after 48 h of incubation.
- Determine the average number of revertants from the two plates used for each selective media at the respective day of incubation; calculate the mutation frequency using the accumulated number of revertants and the number of CFU plated.
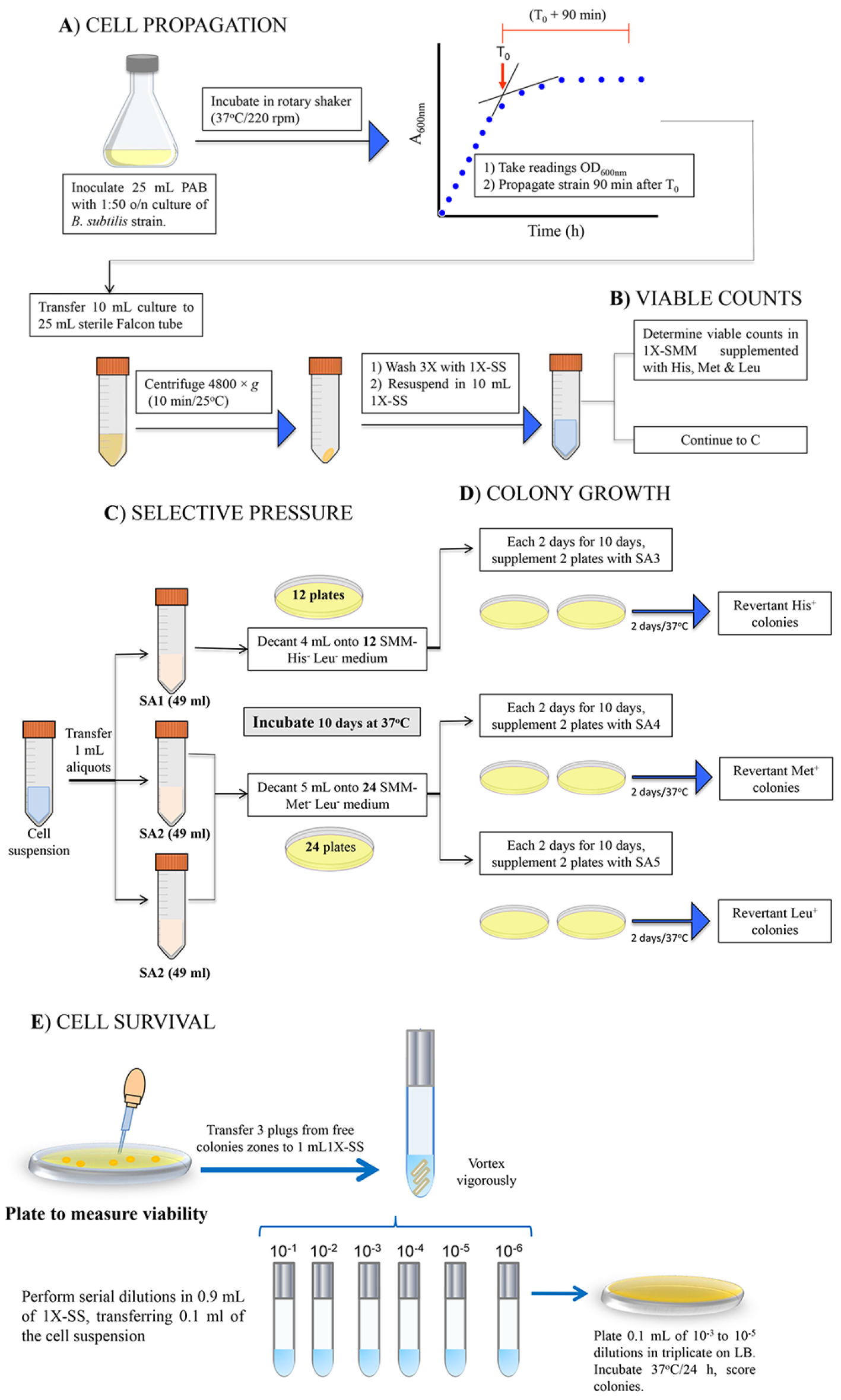
Figure 1. Procedure for Stationary-phase mutagenesis soft-agar overlay assays. A. Cell propagation. Cultivate Bacillus subtilis strains PERM1017, YB955 and PERM1202 in PAB medium until 90 min after the end of exponential growth; wash with 1x SS. B. Viable counts, colony forming units are determined by serial ten-fold dilutions. C. Selective pressure. Cells are plated on selective medium using the corresponding soft agar overlay (SA1 or SA2) and incubated for 10 days at 37 °C. D. Colony growth. A second soft agar overlay (SA3, SA4 or SA5) is added to a set of plates of each selective medium to restore one of the missing amino acids, revertant His+, Met+ and Leu+ colonies are scored after 48 h of incubation. E. Cell survival is monitored during days 2, 4, 6, 8, and 10 from selection SMM-His-Leu- and SMM-Met-Leu- plates as described in Procedure E. In case of strain PERM1017, media must be supplemented with 1 mM IPTG, as indicated in the procedure (perform this procedure on days 2, 4, 6, 8 and 10) (Pybus et al., 2010).
- During days 2, 4, 6, 8, and 10, pick two plates of His- Leu- medium and four plates of Met- Leu-, proceed to prepare soft agar to obtain His- (SA3), Met- (SA4) or Leu- (SA5) media (see Recipes). Over these SMM plates pour a first overlay with the cells and a second overlay with the selective medium (Figure 1 and Supplemental Figure S2). This results in single amino acid dropout media that selects for His+, Met+ or Leu+ revertants. Incubate the plates at 37 °C and record the number of revertants His+, Met+, and Leu+ colonies that arise after 48 h of incubation.
- Cell survival
- Determine the abilities of strains with different genotypes to survive in SMM-His-Leu- or SMM-Met-Leu- plates during the ten days of the experiment.
- Employing sterile Pasteur pipettes, during days, 2, 4, 6, and 8, remove three-agar plugs from areas free of colonies of media plates employed for ‘selective pressure’ (Procedure C). This procedure allows quantifying the initially plated fraction of non-revertant cells that have survived the incubation period.
- Suspend the plugs in 400 µl of 1x SS, vortex thoroughly, plate serial ten-dilutions on SMM containing all the essential amino acids (50 μg/ml). Score the number of colonies after 48 h of incubation at 37 °C.
- Determine the abilities of strains with different genotypes to survive in SMM-His-Leu- or SMM-Met-Leu- plates during the ten days of the experiment.
Data analysis
Following this procedure, Figure 2 shows the accumulation of His+, Met+ and Leu+ revertants in the parental strain YB955 as well as in the conditional RNR strain (PERM1017), under conditions that induce (+ IPTG) or not (- IPTG) the expression of the nrdEF operon.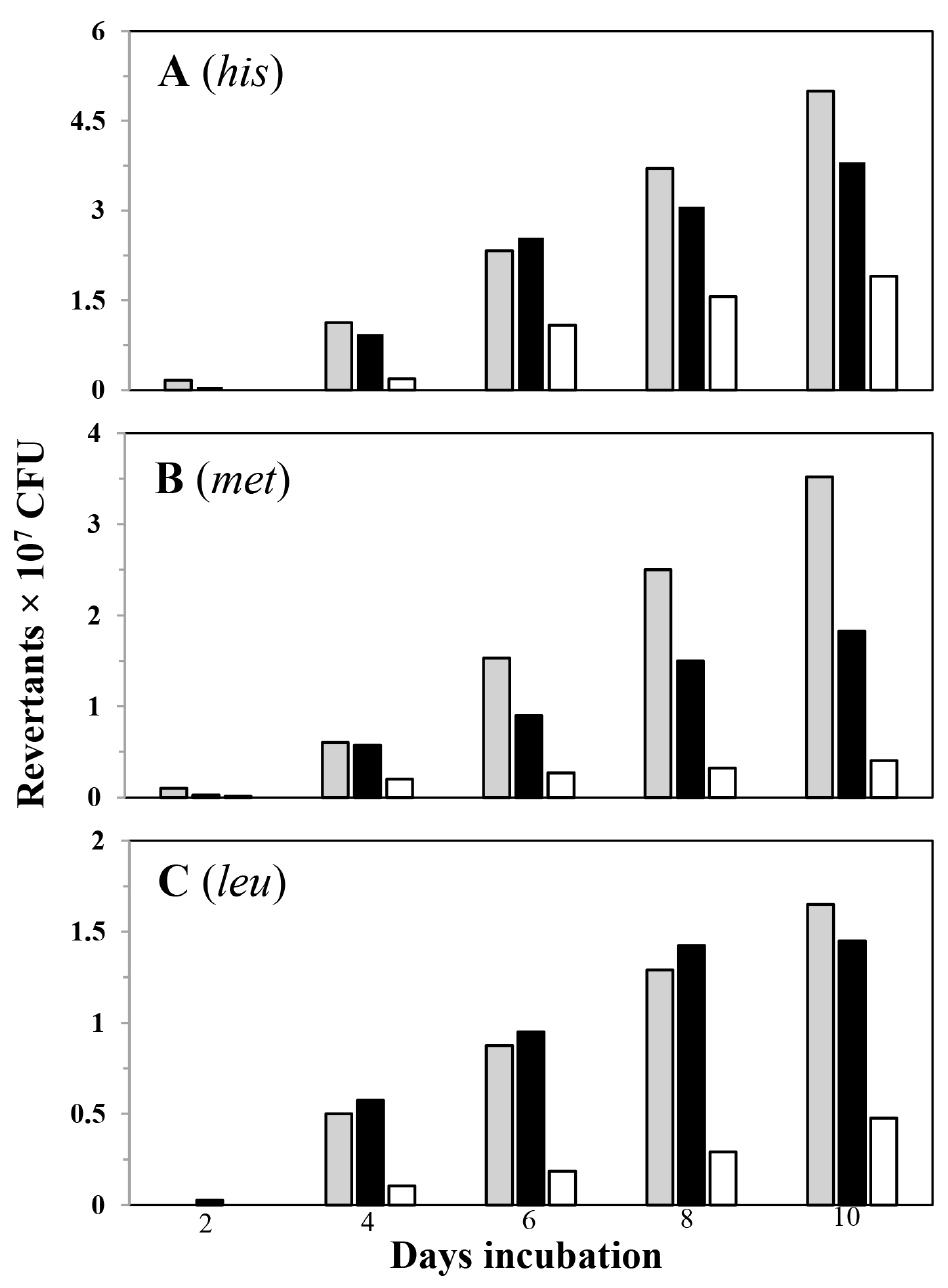
Figure 2. Frequencies of stationary-phase reversions to His+ (A), Met+ (B) and Leu+ (C) of strains B. subtilis YB955 (gray squares), PERM1017 [(conditional RNR strain) (+ IPTG; black squares)] PERM1017 (- IPTG; white squares) were determined as described in procedures. Data represent counts averaged from two separate tests ± standard deviation, Data presented for strains YB955 and PERM1017 (- IPTG) were reported in: Castro-Cerritos, K. V., Yasbin, R. E., Robleto, E. A. and Pedraza-Reyes, M. (2017). Role of ribonucleotide reductase in Bacillus subtilis stress-associated mutagenesis. J Bacteriol 199(4).
Recipes
- PAB medium (PAB)
Dissolve 1.75 g of PAB medium in 100 ml dH2O, autoclave at 121 °C for 15 min - Amino acid solutions [5 mg/ml]
Dissolve 250 mg of the required amino acid in 50 ml of ddH2O
Sterilize by filtration - 1 M IPTG
Dissolve 1.19 g of IPTG in 5 ml of ddH2O and sterilize by filtration - 10x Spizizen salts (10x SS)
Dissolve:
10 g (NH4)2SO4
70 g K2HPO4
30 g KH2PO4
5 g sodium citrate·2H2O
1 g MgSO4·7H2O
Add ddH2O to 500 ml
Autoclave at 121 °C for 15 min - 100x trace elements
Dissolve:
1.28 g MgCl2·6H2O
0.055 g CaCl2
0.135 g FeCl2·6H2O
0.01 g ZnCl2
0.0043 g CuCl2·2H2O
0.006 g CoCl2·6H2O
0.006g Na2MoO4·2H2O
Add ddH2O to 100 ml
Autoclave at 121 °C for 15 min - Spizizen minimal medium (SMM)
In 1 L Erlenmeyer flask:
Add 7.5 g of agar to 430 ml of distilled water, add a magnetic stirring bar
Autoclave at 121 °C for 15 min
Add 50 ml of sterile 10x SS
Cool to 50-55 °C under constant stirring
Add 5 ml of 0.5 g/ml dextrose and a mixture of the following amino acid solutions under constant stirring:- For medium supplementing histidine, methionine, and leucine:
5 ml of 5 mg/ml isoleucine
5 ml of 5 mg/ml glutamic acid
5 ml of 5 mg/ml methionine
5 ml of 5 mg/ml histidine
5 ml of 5 mg/ml leucine - For medium lacking histidine and leucine (SMM-His-Leu-):
5 ml of 5 mg/ml isoleucine
5 ml of 5 mg/ml glutamic acid
5 ml of 5 mg/ml methionine
20 µl of 5 mg/ml histidine
20 µl of 5 mg/ml leucine - For medium lacking methionine and leucine (SMM-Met-Leu-):
5 ml of 5 mg/ml isoleucine
5 ml of 5 mg/ml glutamic acid
5 ml of 5 mg/ml histidine
20 µl of 5 mg/ml methionine
20 µl of 5 mg/ml leucine
Pour:- 4.0 ml of SMM-His-Leu- into 12 (90 x 15 mm) Petri dishes
- 4.0 ml of SMM-Met-Leu- into 24 (90 x 15 mm) Petri dishes
- For medium supplementing histidine, methionine, and leucine:
- Soft agar medium (SMA) for cell inoculation
- 0.7% agar
Add 1.4 g agar to 200 ml of water in a 1 L Erlenmeyer flask with a magnetic stirring bar, autoclave at 121 °C for 15 min.
- For medium lacking histidine and leucine (His- Leu- medium; SA1):
5 ml 10x SS
500 µl of 5 mg/ml isoleucine
500 µl of 5 mg/ml glutamic acid
500 µl of 5 mg/ml methionine
500 µl of 0.5 g/ml dextrose - For medium lacking methionine and leucine (Met- Leu- medium; SA2):
5 ml 10x SS
500 µl 5 mg/ml isoleucine
500 µl 5 mg/ml glutamic acid
500 µl of 5 mg/ml histidine
500 µl 0.5 g/ml dextrose
- Supplement the media with 1 mM IPTG in the case of B. subtilis PERM1017.
- Add pre-warmed agar (50-55 °C) to a final volume of 50 ml in each tube; mix gently.
- 0.7% agar
- Soft agar to obtain His-, Met- or Leu- media
- Add 350 mg agar to 50 ml of water in a flask with a magnetic stirrer bar, autoclave at 121 °C for 15 min.
- For His- medium (SA3):
1 ml of 10x SS
1 ml of 5 mg/ml leucine
100 µl of 5 mg/ml isoleucine
100 µl of 5 mg/ml glutamic acid
100 µl of 5 mg/ml methionine
100 µl of 0.5 g/ml dextrose - For Met- medium (SA4):
1 ml of 10x SS
1 ml of 5 mg/ml histidine
100 µl of 5 mg/ml isoleucine
100 µl of 5 mg/ml glutamic acid
100 µl of 5 mg/ml leucine
100 µl of 0.5 g/ml dextrose - For Leu- medium (SA5):
1 ml 10x SS
1 ml of 5 mg/ml methionine
100 µl of 5 mg/ml isoleucine
100 µl of 5 mg/ml glutamic acid
100 µl of 5 mg/ml histidine
100 µl of 0.5 g/ml dextrose
- In the case of B. subtilis PERM1017, 100 µl of 1 M IPTG must be added to the mixtures to reach a final concentration of 1 mM (considering a final volume of 50 ml in the Petri dish).
- Add pre-warmed agar (50-55 °C) to a final volume of 10 ml in each tube; mix gently.
- Add 350 mg agar to 50 ml of water in a flask with a magnetic stirrer bar, autoclave at 121 °C for 15 min.
Acknowledgments
This work was supported by the Consejo Nacional de Ciencia y Tecnología (CONACYT; Grants 205744 and 221231) of México and by the University of Guanajuato (Grants 936-2016 and 1090-2016) to M.P-R. Work at ER was supported by the NIH grant (R15GM110624). KV. C-C and N. V-N were supported by scholarships from CONACYT. The authors declare no conflict of interest.
References
- Barajas-Ornelas R del, C., Ramirez-Guadiana, F. H., Juarez-Godinez, R., Ayala-Garcia, V. M., Robleto, E. A., Yasbin, R. E. and Pedraza-Reyes, M. (2014). Error-prone processing of apurinic/apyrimidinic (AP) sites by PolX underlies a novel mechanism that promotes adaptive mutagenesis in Bacillus subtilis. J Bacteriol 196(16): 3012-3022.
- Castro-Cerritos, K. V., Yasbin, R. E., Robleto, E. A. and Pedraza-Reyes, M. (2017). Role of ribonucleotide reductase in Bacillus subtilis stress-associated mutagenesis. J Bacteriol 199(4).
- Gómez-Marroquín, M., Martin, H. A., Pepper, A., Girard, M. E., Kidman, A. A., Vallin, C., Yasbin, R. E., Pedraza-Reyes, M. and Robleto, E. A. (2016). Stationary-phase mutagenesis in stressed Bacillus subtilis cells operates by Mfd-dependent mutagenic pathways. Genes (Basel) 7(7).
- Kobayashi, K., Ehrlich, S. D., Albertini, A., Amati, G., Andersen, K. K., Arnaud, M., Asai, K., Ashikaga, S., Aymerich, S., Bessieres, P., Boland, F., Brignell, S. C., Bron, S., Bunai, K., Chapuis, J., Christiansen, L. C., Danchin, A., Debarbouille, M., Dervyn, E., Deuerling, E., Devine, K., Devine, S. K., Dreesen, O., Errington, J., Fillinger, S., Foster, S. J., Fujita, Y., Galizzi, A., Gardan, R., Eschevins, C., Fukushima, T., Haga, K., Harwood, C. R., Hecker, M., Hosoya, D., Hullo, M. F., Kakeshita, H., Karamata, D., Kasahara, Y., Kawamura, F., Koga, K., Koski, P., Kuwana, R., Imamura, D., Ishimaru, M., Ishikawa, S., Ishio, I., Le Coq, D., Masson, A., Mauel, C., Meima, R., Mellado, R. P., Moir, A., Moriya, S., Nagakawa, E., Nanamiya, H., Nakai, S., Nygaard, P., Ogura, M., Ohanan, T., O'Reilly, M., O'Rourke, M., Pragai, Z., Pooley, H. M., Rapoport, G., Rawlins, J. P., Rivas, L. A., Rivolta, C., Sadaie, A., Sadaie, Y., Sarvas, M., Sato, T., Saxild, H. H., Scanlan, E., Schumann, W., Seegers, J. F., Sekiguchi, J., Sekowska, A., Seror, S. J., Simon, M., Stragier, P., Studer, R., Takamatsu, H., Tanaka, T., Takeuchi, M., Thomaides, H. B., Vagner, V., van Dijl, J. M., Watabe, K., Wipat, A., Yamamoto, H., Yamamoto, M., Yamamoto, Y., Yamane, K., Yata, K., Yoshida, K., Yoshikawa, H., Zuber, U. and Ogasawara, N. (2003). Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A 100(8): 4678-4683.
- Pybus, C., Pedraza-Reyes, M., Ross, C. A., Martin, H., Ona, K., Yasbin, R. E. and Robleto, E. (2010). Transcription-associated mutation in Bacillus subtilis cells under stress. J Bacteriol 192(13): 3321-3328.
- Sung, H. M. and Yasbin, R. E. (2002). Adaptive, or stationary-phase, mutagenesis, a component of bacterial differentiation in Bacillus subtilis. J Bacteriol 184(20): 5641-5653.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Castro-Cerritos, K. V., Villegas-Negrete, N., Ramirez-Ramirez, N., Robleto, E. A. and Pedraza-Reyes, M. (2017). Stationary-phase Mutagenesis Soft-agar Overlay Assays in Bacillus subtilis. Bio-protocol 7(23): e2634. DOI: 10.21769/BioProtoc.2634.
Category
Microbiology > Microbial genetics > Mutagenesis
Molecular Biology > DNA > Mutagenesis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link


.jpg)

.jpg)







