- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Flow-assay for Farnesol Removal from Adherent Candida albicans Cultures
Published: Vol 7, Iss 19, Oct 5, 2017 DOI: 10.21769/BioProtoc.2562 Views: 8926
Reviewed by: Valentine V TrotterEmmanuel ZavalzaJose Thekkiniath

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Bioassay Protocol for Quorum Sensing Studies Using Vibrio campbellii
Alberto J. Martín-Rodríguez and José J. Fernández
Jul 20, 2016 11813 Views

Protein Expression Protocol for an Adenylate Cyclase Anchored by a Vibrio Quorum Sensing Receptor
Stephanie Beltz and Joachim E. Schultz
Jan 20, 2017 9387 Views

Extraction of Small Molecules from Fecal Samples and Testing of Their Activity on Microbial Physiology
Eduardo S. Alves [...] L. Caetano M. Antunes
Apr 20, 2018 7987 Views
Abstract
Here, we describe a protocol for a continuous flow system for C. albicans cultures growing adherent to a plastic surface. The protocol was adapted from a previous method established to simulate blood flow on endothelial cells (Wilson and Hube, 2010). The adapted protocol was used by us for the removal of molecules in C. albicans supernatants, especially farnesol, which accumulate over the time course of incubation and cannot be specifically depleted. The system used, however, allows various applications including the simulation of physiological flow conditions. Several example applications are given on the manufacturer’s website (https://ibidi.com/perfusion-system/112-ibidi-pump-system.html).
Keywords: Continuous flowBackground
Farnesol is a potent inhibitor of the yeast-to hypha transition (Hornby et al., 2001) in the human pathogenic fungus Candida albicans and also promotes the reversal to yeast growth from preformed filaments (Lindsay et al., 2012). The quorum sensing molecule (QSM) rapidly accumulates in the supernatant of a Candida albicans EED1 deletion strain and promotes the reverse morphogenesis and a hyphal maintenance defect of the mutant (Polke et al., 2017). As we were unable to block farnesol synthesis (Polke et al., 2017), we utilized the ibidi® pump system to remove the accumulating QSMs in the supernatant by uni-directional flow. Flow application, together with a constant medium exchange during the time course of incubation, significantly prolonged filamentation in the C. albicans eed1∆ mutant. This indicated the successful removal of QSM accumulates, and provided a direct link between hyphal maintenance and farnesol signaling in C. albicans. The system used for this protocol (ibidi® pump system) allows various applications under simulation of physiological flow conditions, and thus might be easily modified for other applications. Several example applications are given on the manufacturer’s website (https://ibidi.com/perfusion-system/112-ibidi-pump-system.html).
Materials and Reagents
- Protective gloves and lab coat
- Pipette tips (TipOne) (STARLAB INTERNATIONAL, catalog numbers: S1111-6000 , S1113-1006 , S1110-3000 )
- µ-Slide VI0.4 ibiTreat: #1.5 polymer coverslip, tissue culture treated, sterilized (ibidi, catalog number: 80606 )
- Flask with absorber beads: dry beads (KC Trockenperlen® [Sorbead®] orange, BASF)
- Micro-tubes, 1.5 ml (SARSTEDT, catalog number: 72.690.001 )
- 0.2 μm sterile filters (Minisart 0.2) (SARSTEDT, catalog number: 83.1862.001 )
- Syringe Injekt® 10 ml/Luer Lock Solo, sterile (B. Braun Medical, catalog number: 4606728V-02 )
- 50 ml Falcon tubes (SARSTEDT, catalog number: 62.547.254 )
- 10 ml pipette, graduated, sterile (Greiner Bio One International, catalog number: 607180 )
- Petri dishes (Greiner Bio One International, catalog number: 633180 )
- Disposal bags (Carl Roth, catalog number: E706.1 )
- Steam Indicator Tape 3M (ComplyTM, 3M, catalog number: 1322-18MM )
- Perfusion Set Blue (ibidi, catalog number: 10961 )
- Filter/Reservoir set (10 ml, sterile) (ibidi, catalog number: 10971 )
- Candida albicans strains of interest (the system was established using SC5314 and the respective EED1 deletion mutant, see Polke et al., 2017)
- RPMI1640 medium [(+)L-glutamine, (+)phenol red, unbuffered] (Thermo Fisher Scientific, GibcoTM, catalog number: 21875034 )
- Fermacidal D2® (2%) (LABOTECT, catalog number: 15101 )
- Roti®-Histofix 4% (Carl Roth, catalog number: P087.5 )
- Glycerol, ROTIPURAN®, water-free (Carl Roth, catalog number: 3783.2 )
- D(+)-Glucose, water-free (Carl Roth, catalog number: HN06.4 )
- BactoTM peptone (BD, BactoTM, catalog number: 211677 )
- Yeast extract, micro-granulated (Carl Roth, catalog number: 2904.1 )
- Agar-agar, Kobe I (Carl Roth, catalog number: 5210.4 )
- Sodium chloride (NaCl) (Carl Roth, catalog number: 9265.2 )
- Disodium phosphate (Na2HPO4·2H2O) (Carl Roth, catalog number: T877.1 )
- Monopotassium phosphate (KH2PO4) (Carl Roth, catalog number: 3904.1 )
- Ethanol denatured ≥ 99.8% (Carl Roth, catalog number: K928.4 )
- 30% glycerin solution (see Recipes)
- 20% D(+)-glucose solution (see Recipes)
- YPD broth (see Recipes)
- YPD agar medium (see Recipes)
- 10x PBS (see Recipes)
- 1x PBS (see Recipes)
- 70% ethanol (see Recipes)
Equipment
- Milli-Q® integral water purification system for ultrapure water (deionized water) (Merck, model: Milli-Q® Integral )
- Infors HT, Multitron Standard shaking incubator, Version 2 (Infors, model: Multitron Standard )
- BINDER cooling incubator (series: APT.line®KB, BINDER, model: KB 53 ; 30 °C)
- BINDER CO2 incubator (series APT.line®CB, BINDER, model: CB 220 ; 37 °C)
- Glass flasks 25 ml and 250 ml (Schott, DURAN, Germany)
- Pipette set 0.2 μl-1,000 μl (Gilson, model: PIPETMAN® P, P2 , P10 , P20 , P100 , P200 and P1000 )
- Tabletop centrifuge (Thermo Fisher Scientific, Thermo ScientificTM, model: HereausTM PicoTM 21 )
- Mid bench centrifuge (Sigma Laborzentrifugen, model: SIGMA 3-18K )
- Vortexer (Scientific Industries, model: Vortex-Genie 2 )
- Neubauer improved, cell counting chamber 0.0025 mm2 (Marienfeld-Superior, catalog number: 0640030 )
- Biosafety cabinet (NuAire, model: NU-480-400E )
- Ibidi® pump system including ibidi pump, fluidic unit, perfusion set, notebook, PumpControl software (ibidi, catalog number: 10902 )
- ZEISS inverted microscope (ZEISS, model: Axio Vert.A1 )
- UV Crosslinker (Vilber, model: Bio-Link 254 )
- Autoclave (for example: SHP Steriltechnik, model: Laboklav 135 MSLV )
Software
- Computer equipped with ZEISS ZEN software (Blue edition, 2012)
- Computer with GraphPad Prism 5 software
Procedure
- Growth of Candida albicans cells and germ tube induction
- Streak respective C. albicans strains from 30% glycerol stocks (see Recipes) onto YPD agar (see Recipes) and grow for 2 d at 30 °C in an incubator.
- Pick a single colony from each strain and re-streak onto a fresh YPD agar plate. Grow for 2 d at 30 °C in an incubator.
- Pick a single colony from each strain and inoculate in 10 ml YPD medium (see Recipes) in a 25 ml glass flask. Grow overnight at 30 °C with vigorous horizontal shaking (180 rpm) in a shaking incubator.
- Dilute the o/n culture 100-fold (100 μl) in fresh 10 ml YPD medium and grow overnight (approximately 20 h) in a 25 ml glass flask at 30 °C with horizontal shaking (180 rpm) to semi-synchronize the growth of all strains.
- On the day of the experiment harvest 1 ml of the overnight culture of each strain into a 1.5 ml micro-tube and centrifuge at 10,000 x g for 1 min at room temperature in a tabletop centrifuge to collect cells. Remove the supernatant and resuspend the cells in 1 ml sterile PBS (see Recipes) to wash the yeast.
- Repeat the washing step described above twice.
- After the last wash, resuspend the yeast in 1 ml PBS.
- Prepare an appropriate dilution of cells for determination of the total cell number in suspension; for the wild type C. albicans strain, a 20 h culture prepared as described above, a 100-fold dilution of the initial culture is usually appropriate.
- Apply 10 μl of the dilution onto a Neubauer cell counting chamber and determine the cell number per ml using a microscope following the manufacturer’s instructions.
- Seed 1 x 105 cells/ml into 50 ml RPMI1640 medium (pre-warmed at 37 °C) in a 250 ml glass flask and incubate with shaking (180 rpm) for 30 min at 37 °C to induce germ tube formation.
- Streak respective C. albicans strains from 30% glycerol stocks (see Recipes) onto YPD agar (see Recipes) and grow for 2 d at 30 °C in an incubator.
- Equilibrate the RPMI1640 medium and the materials (perfusion set) before the experiment at 37 °C and 5% CO2 (incubation conditions during the flow assay); this is best performed overnight.
Note: Prepare the equipment of the ibidi® pump system and the incubation setup before harvesting the C. albicans cultures to be ready to start when the germ tubes are formed (see above). The preparation of the fluidic unit and the perfusion set must occur under sterile conditions. The µ-slide must be prepared and connected to the pump system before the complete unit can be placed in the incubator. The assembly of the system must be performed in a biosafety cabinet. - Prepare the perfusion set (see Figure 1)
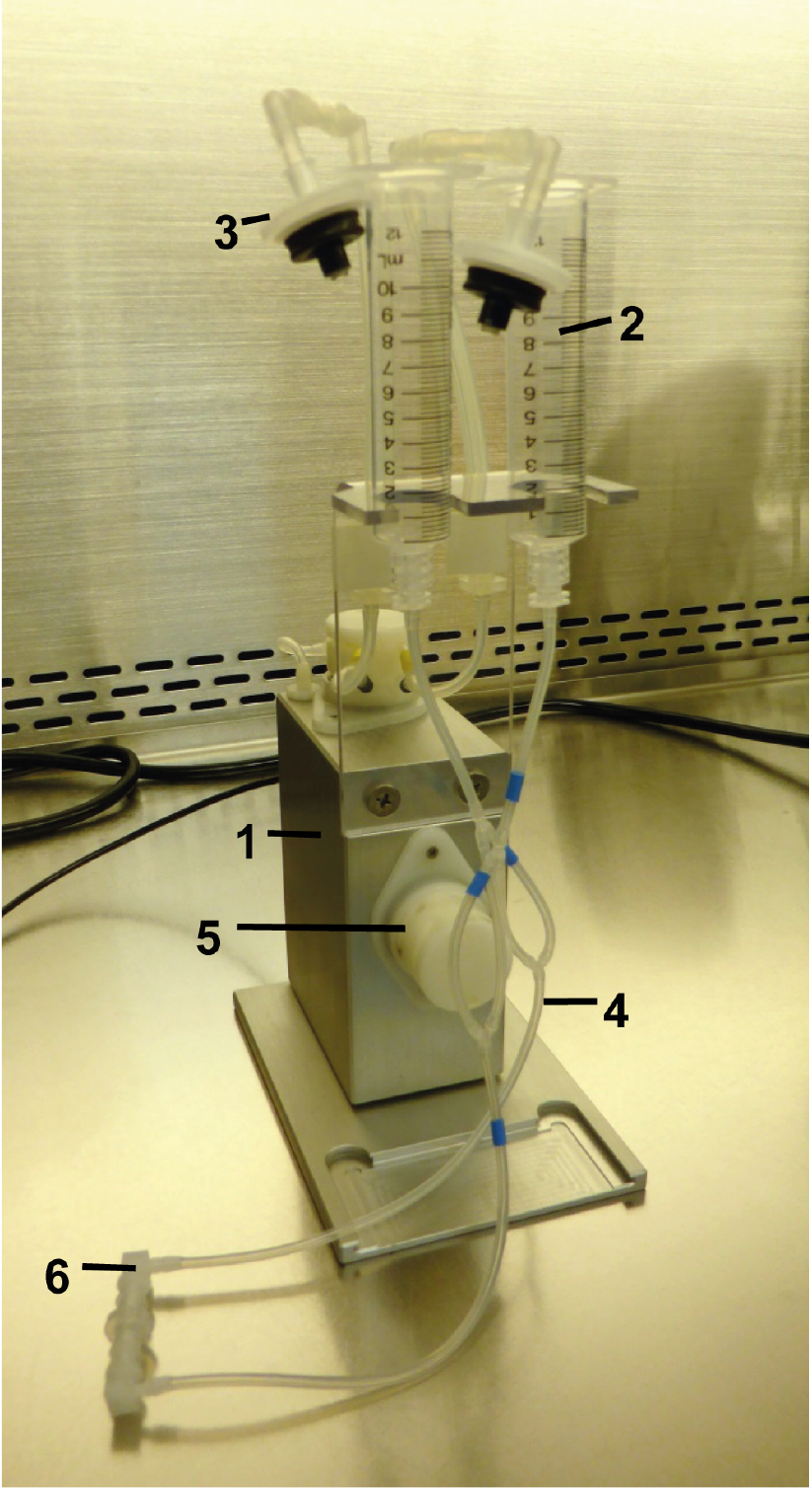
Figure 1. Fluidic unit with perfusion set. The two reservoirs have been mounted to the holder of the fluidic unit. Prepare the unit under a clean bench for sterility. Fluidic unit (1), reservoir 10 ml (2), 0.2 μm sterile filter (3), tubing (4), valve (5) and tube-endings with Luer locks connected by the Luer Lock Coupler (6).- Attach the reservoirs of the perfusion set to the fluidic unit; close/connect the ends of the tubings with the included Luer Lock Coupler, and remove the mounted filters capping the reservoirs.
- Add ~10 ml of equilibrated RPMI1640 medium to each of the reservoirs (Figure 2A). The medium will spread across the tubing system.
- To remove remaining air bubbles in the tubes, open the Luer Lock Coupler connection (medium will leak from the tube so make sure to collect it in a Falcon tube or to soak it on a paper sheet). Use a sterile plug from a 10 ml syringe to apply pressure on the medium by inserting the plug into the perfusion set reservoirs. Remove all air bubbles by pushing medium through the system (Figures 2B and 2C). Avoid letting the tubing system run dry as this will cause new air bubbles to arise.
- When all air bubbles are removed, sterilize the Luer connection at both ends with 2% Fermacidal (or another suitable disinfectant) and reconnect the ends of the tubings with the Luer Lock Coupler. Fill up each reservoir with equilibrated RPMI1640 medium to 5-5.5 ml (Figure 2D; the total working volume of the Blue Perfusion set system is 11.3 ml).
- Close the reservoirs by mounting the filters (Figure 2E). The filters are necessary to avoid contamination with air-borne microbes through the tubing system during pressure application by the pump.
- Place the prepared fluidic unit, including the prepared perfusion set, into the CO2 incubator (37 °C 5% CO2) for pre-warming until the beginning of the experiment (for at least 1 h if the media was pre-warmed; the unit can be set up the day before the actual experiment).
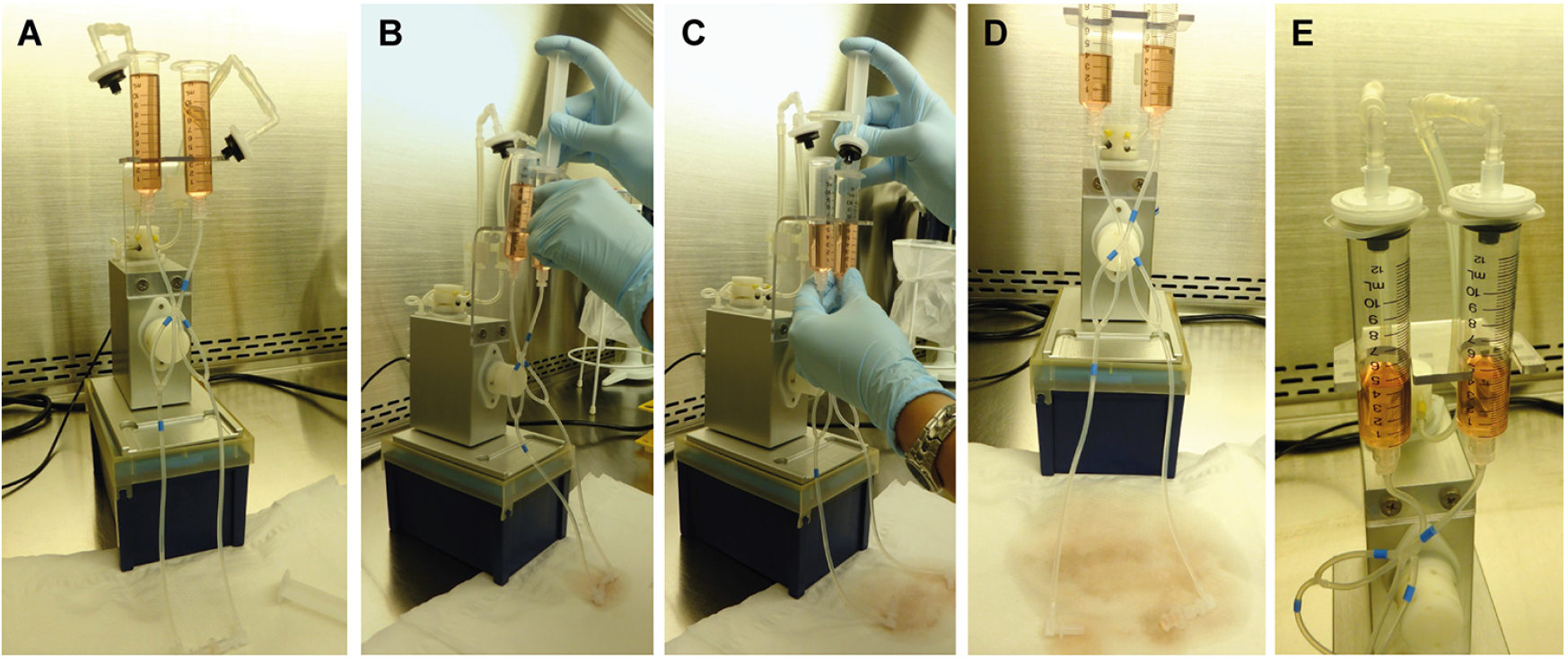
Figure 2. Preparation of the fluidic unit and removal of air bubbles in the system. Fill both reservoirs with medium (A). The medium will spread through the tubings, but air bubbles remain. To remove air bubbles, use a 10 ml syringe plunge to apply weak pressure in both reservoirs (B and C). Make sure to collect leaking (sterile) medium in a tube or on paper sheets (D). When all air bubbles are removed, disinfect the Luer connector and tube endings, reconnect the tube endings with the Luer Lock Coupler, and close the reservoirs with the supplied filters.
- Attach the reservoirs of the perfusion set to the fluidic unit; close/connect the ends of the tubings with the included Luer Lock Coupler, and remove the mounted filters capping the reservoirs.
- Set up the pump by connecting it to the fluidic unit, power supply, and computer and adjust the settings in the ibidi® pump system software:
- Connect the pump to the notebook and connect all tubings as indicated in Figure 3. Note that a flask with absorber beads must be placed between the pump and the fluidic unit to remove the moisture generated in the tubings during incubation in the humidified incubator chamber.
- Start the software for the ibidi® pump.
- For uni-directional flow with 2 ml/min flow rate choose the parameters as given in Figure 4.
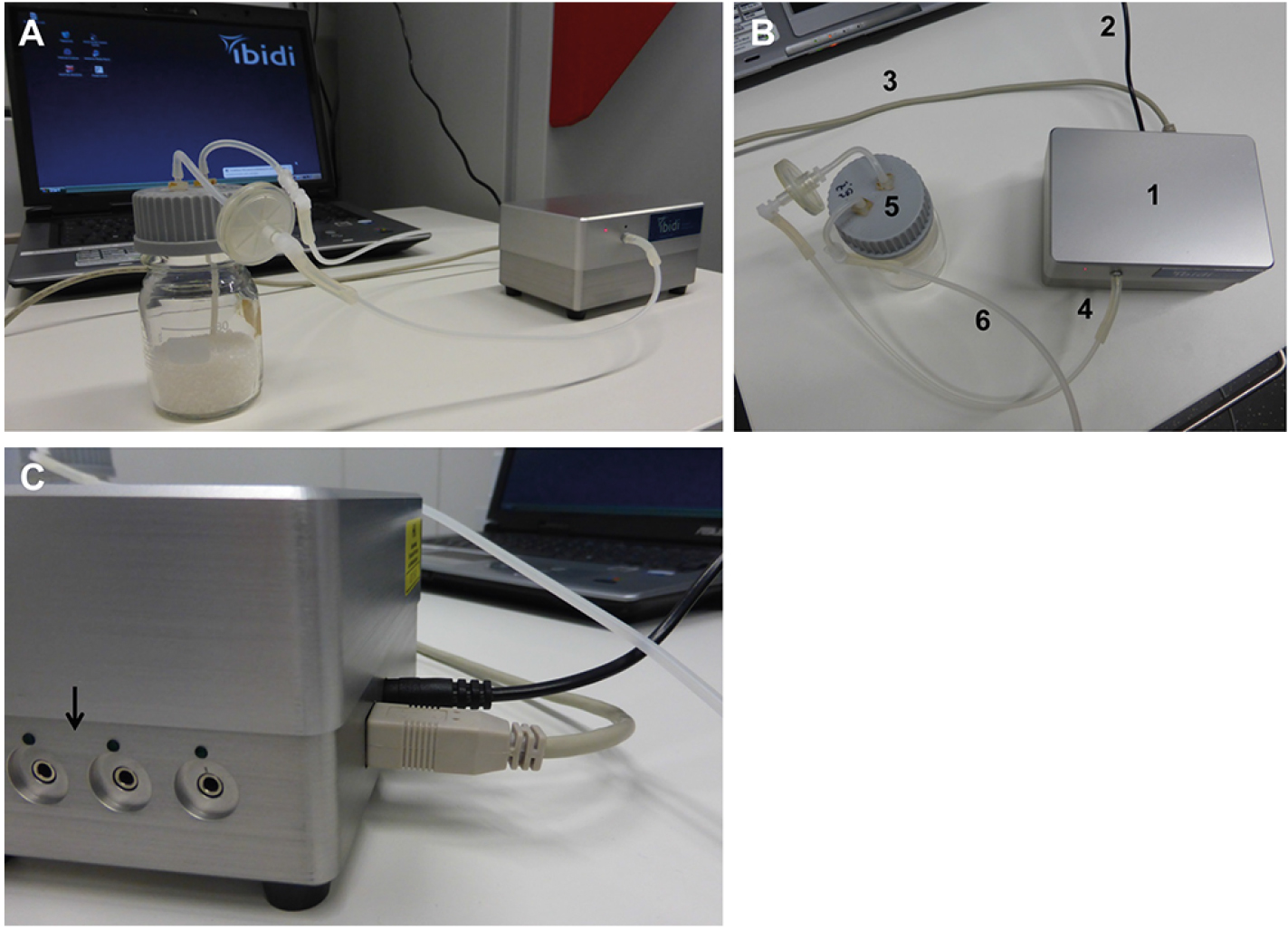
Figure 3. Setup of the computer and pump system. The parts of the system shown in (A) remain on the bench, while the fluidic unit is placed within the CO2 incubator. The connections between the pump, absorber, and computer are indicated in B: The pump (1) is connected to the power supply (2) and the computer (3). The tubing for applying pressure is mounted on the pump (4), and connected to the absorber bottle (5). From the absorber bottle, the long tube (6) will be connected with the fluidic unit in the incubator to apply pressure on the medium in the reservoirs. The pump contains several channels for connection to fluidic units (C, indicated by arrow). Up to four fluidic units can be controlled by one pump. Choose the right connection when setting up the experiment in the software (see Figure 4).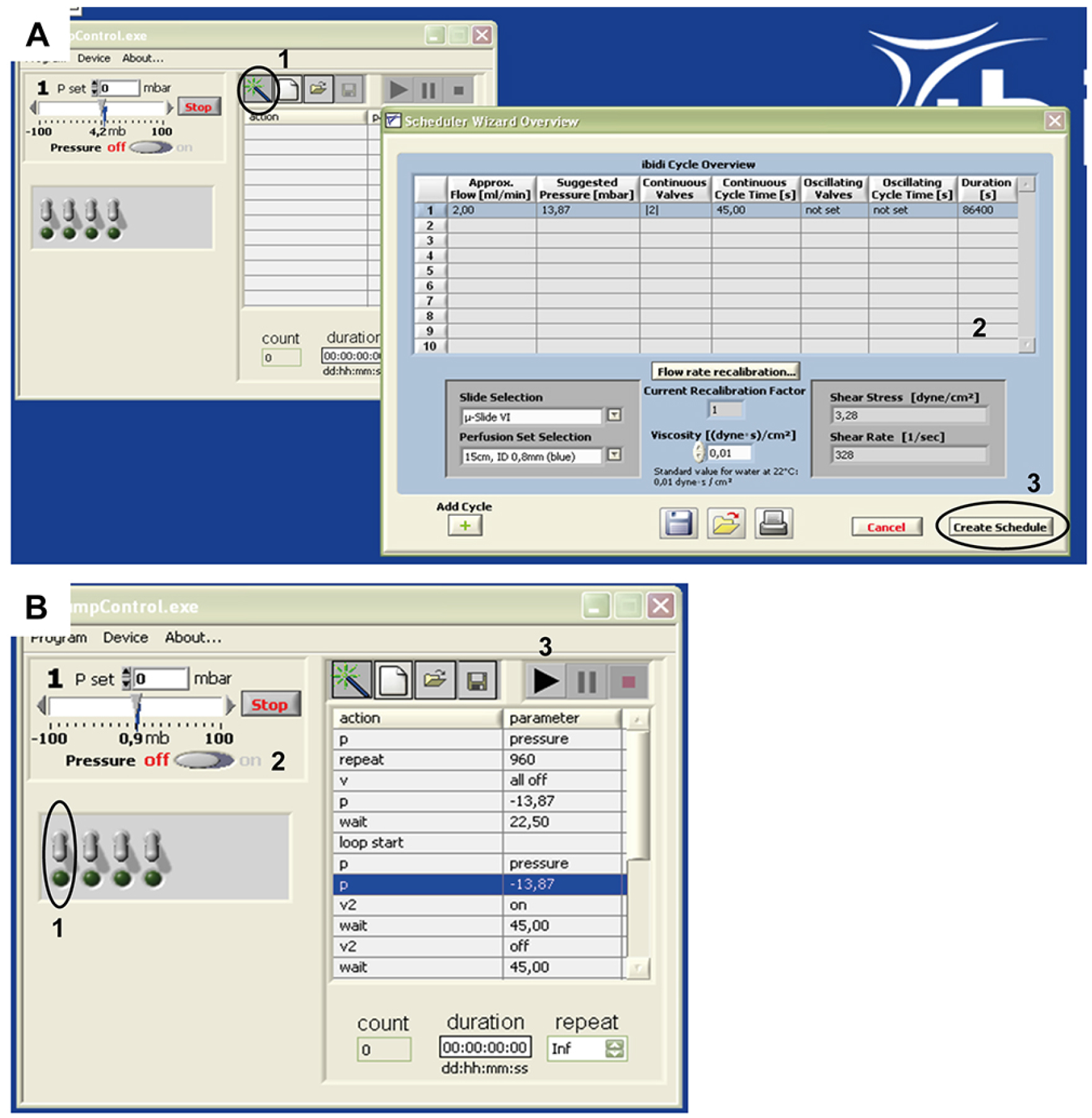
Figure 4. ibidi® pump control software setup for a flow rate of 2 ml/min. A. Open the software and open the Scheduler Wizard Overview by clicking on the button indicated in A1. A new window will open. In our application (Polke et al., 2017), the parameters as indicated in A2 were used: Approx. Flow 2,00 ml/min; Suggested Pressure 13,87 mbar; Continuous Valves ‘2’; Continuous Cycle Time 45,00 sec; Oscillating Valves: ‘not set’; Oscillating Cycle Time: ‘not set’; Duration: 86,400 sec. Slide Selection: µ-slide VI0.4; Perfusion Set Selection: 15 cm, ID 0.8 mm (blue); Viscosity 0,01. When you finished setting up the parameters, select ‘Create Schedule’ as indicated by A3. B. The parameters will appear in the Pump Control; at this point, software and pump are ready for use. To connect the fluidic unit/perfusion to the pump (following step 7 of this protocol), choose the appropriate pump channel (B1, also see Figure 3), apply pressure (B2) and start the flow (B3).
- Connect the pump to the notebook and connect all tubings as indicated in Figure 3. Note that a flask with absorber beads must be placed between the pump and the fluidic unit to remove the moisture generated in the tubings during incubation in the humidified incubator chamber.
- Collect the C. albicans germ tubes and seed onto ibidi μ-slide
- To collect germ tubes, centrifuge the culture prepared according to step 1j in a 50 ml Falcon tube at 4,000 x g for 10 min to 20 min at room temperature. While collection of germ tubes can be tricky and sometimes requires long centrifugation steps, in our hands the germ tubes at 30 min post hypha-induction can be successfully collected by centrifugation at 4,000 x g for 10 min at room temperature.
- Carefully remove the supernatant by aspirating using a pipette (as the cell pellet may be ‘fluffy’ it easily gets lost when the supernatant is decanted). Resuspend cells in 10 ml fresh (pre-warmed to 37 °C) RPMI1640 to get a final cell density of 5 x 105 cells/ml. For our experiments cell density was not critical and we therefore assumed that no cells were lost during supernatant removal; for other applications, however, it might be necessary to re-count the cells before final resuspension to obtain a more accurate cell density.
- Carefully vortex the cell suspension and seed the C. albicans cells on the ibidi μ-slide VI0.4 by adding 30 μl to each channel to be used (e.g., one for flow condition, one for static condition; see Figure 5). The final cell number in the channel (0.6 cm2 of growth area) is 1.5 x 104 cells/ml.
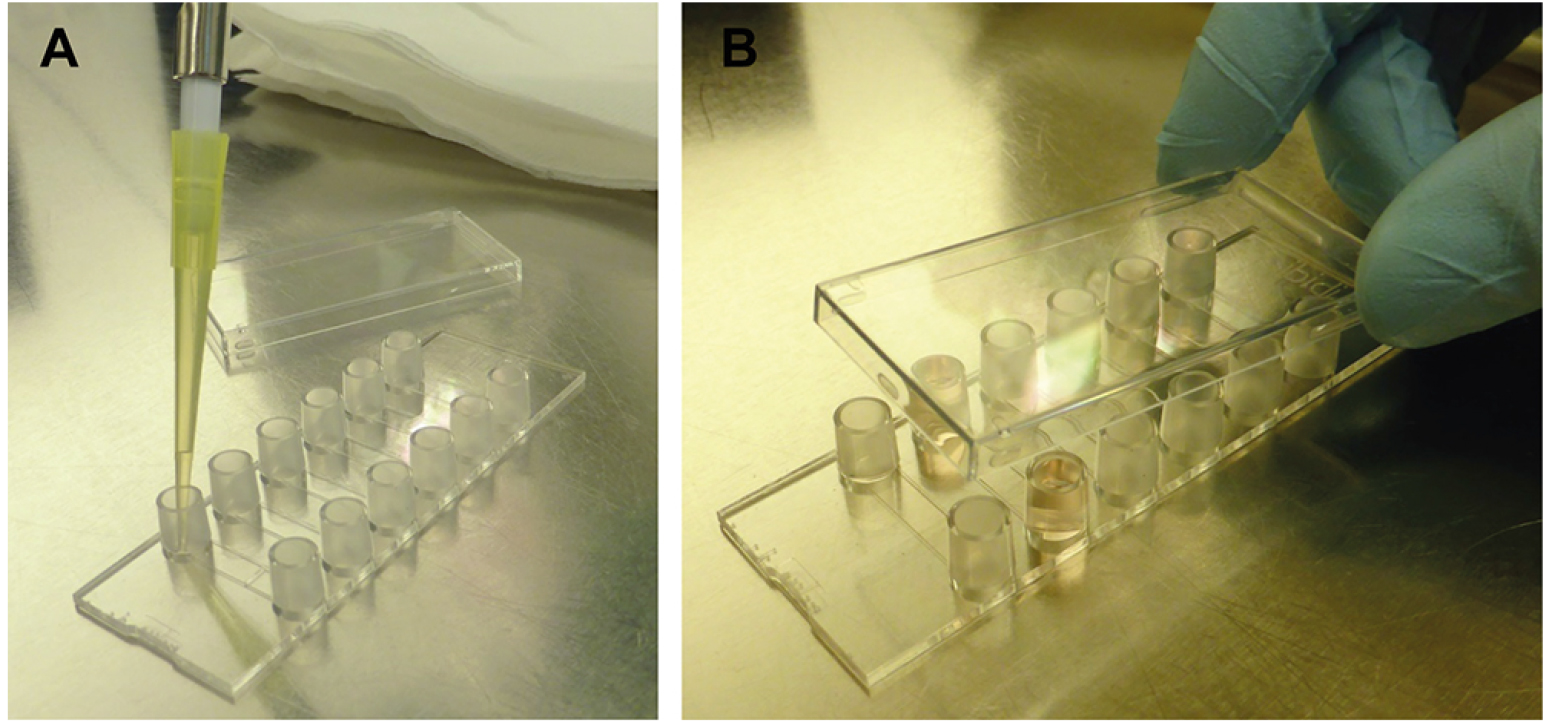
Figure 5. Cell seeding into the ibidi μ-slide VI0.4. Carefully pipette the cell suspension into the small reservoirs of the μ-slide under sterile conditions (A). 30 μl is enough to fill the cell channel, if you wish you can also apply more medium. Close the lid (B), and place the μ-slide in an incubator (37 °C 5% CO2 humidified chamber) for 20 min to allow adherence of cells. - Close the channels with the included lid and incubate for 20 min at 37 °C and 5% CO2 to allow adhesion of germ tubes to the plastic surface. During the flow, all non-adherent cells would be washed off and accumulate in the medium in the reservoirs.
- To collect germ tubes, centrifuge the culture prepared according to step 1j in a 50 ml Falcon tube at 4,000 x g for 10 min to 20 min at room temperature. While collection of germ tubes can be tricky and sometimes requires long centrifugation steps, in our hands the germ tubes at 30 min post hypha-induction can be successfully collected by centrifugation at 4,000 x g for 10 min at room temperature.
- Wash cells and connect ibidi μ-slide to the perfusion set
- Following adhesion, carefully remove the medium from the channels, wash cells once by adding 150 μl pre-warmed RPMI1640 medium and aspirating again. This will remove most non-adherent cells. Avoid vigorous pipetting as this might detach cells.
- For the cells grown under static conditions, apply 120 μl pre-warmed RPMI1640 medium and close the channel with the lid.
- For cells grown under flow conditions, add ~200 μl pre-warmed RPMI1640 medium to the appropriate channel (the channel should be completely filled with fluid [according to Figure 6A]).
- Remove the fluidic unit, including the perfusion set, from the incubator, and place it under a clean bench. Open the Luer Lock Coupler and rapidly attach the Luer connections to the small reservoirs on each end of the channel (see Figure 6). Avoid medium spilling from the tubes. This is easily done when the tubing is temporarily closed with a clip, as visible in Figure 6. The ibidi μ-slide is now prepared for incubation, and can be fixed on the fluidic unit as indicated in Figure 6D.
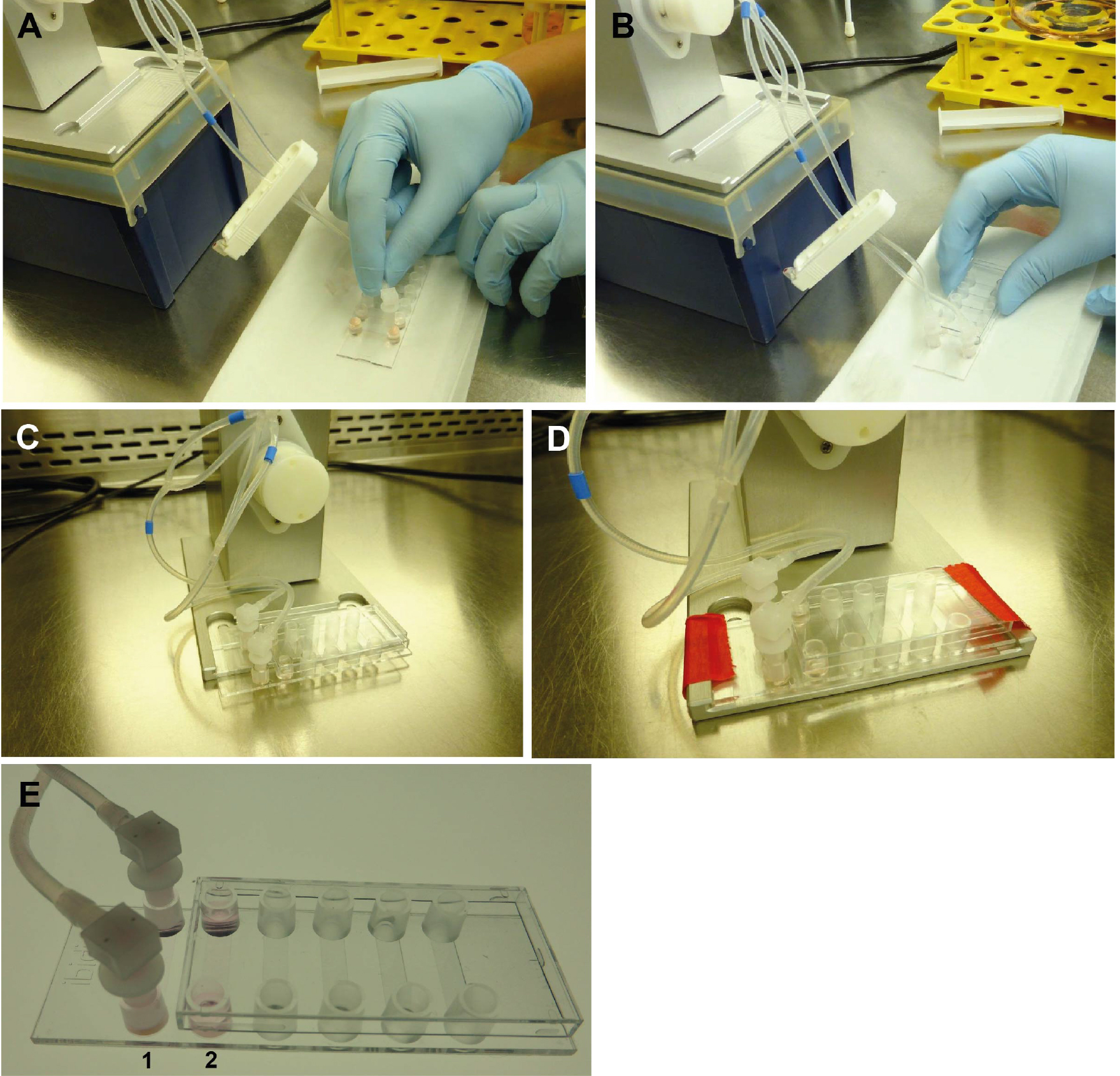
Figure 6. Connecting the ibidi μ-slide VI0.4 to the perfusion set (under sterile conditions). Close the tubes with a standard clip to avoid medium leakage from the tubes during connection. Remove the Luer Lock Coupler and attach each end of the tubes to one of the small reservoirs (A). Make sure that the channel is completely full of fluid to avoid introducing air bubbles into system. Cover the cells grown under static conditions (B). Place the μ-slide on the designated space on the fluidic unit (C) and fix the slide with adhesive tape to avoid movement or slipping during transport (D). Cells are then grown under flow (E1) or static conditions (E2), respectively. - Connect the tubes of the Perfusion set in correct arrangement with the valve set: During the run/flow, medium from one reservoir is pumped through the tubes and the μ-slide channel into the other reservoir and vice versa. In order to allow uni-directional flow in the channel, the fluidic unit contains two different valves which are switched upon pumping in one or the other direction. A nice description of the principle is given on the manufacturer’s website (https://ibidi.com/perfusion-system/112-ibidi-pump-system.html) and in (Wilson and Hube, 2010). To allow the correct switching of valves, the tubes have to be fixed in a criss-crossed order (see Figures 7A and 8).
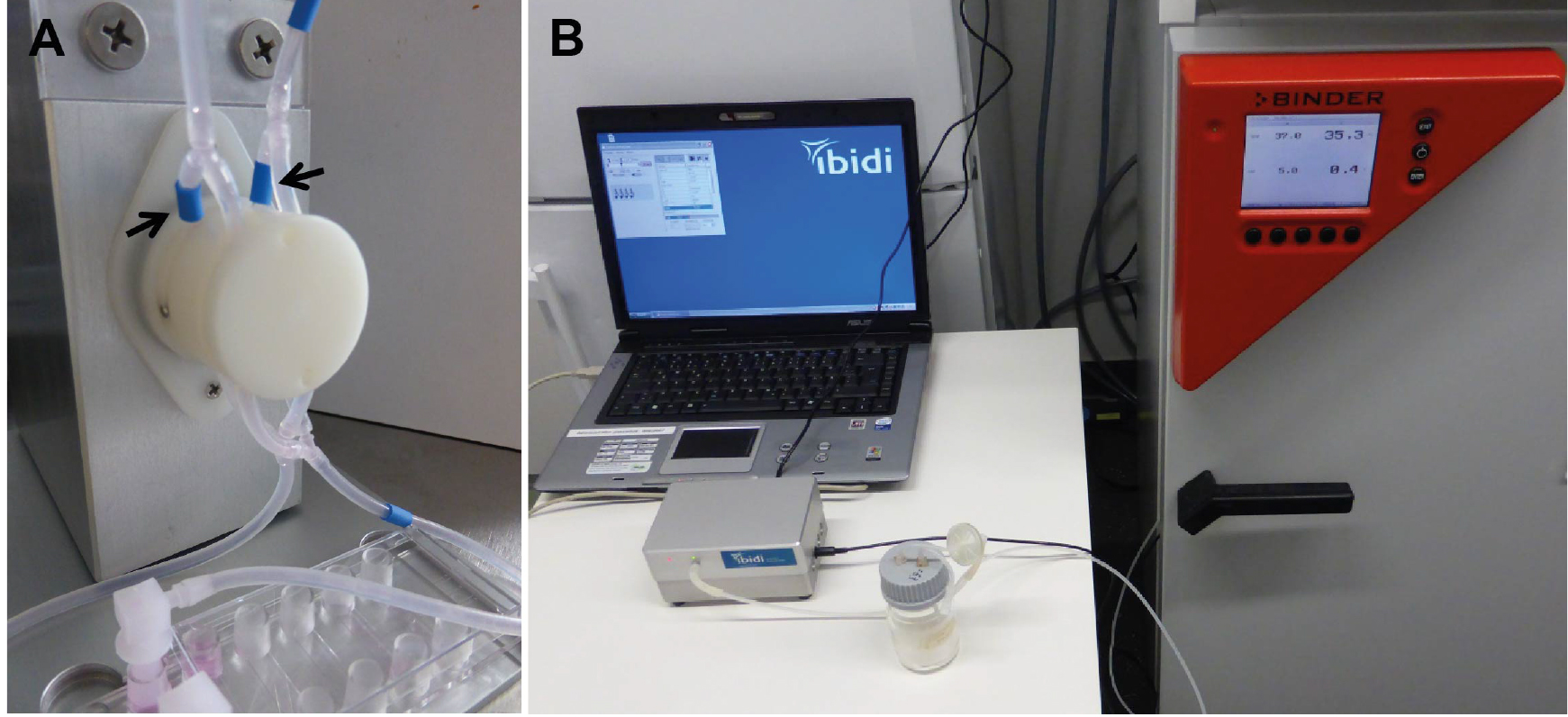
Figure 7. Flow application. Connect the tubings with the valve (A). Make sure to place them in the right order to allow a uni-directional flow in the μ-channel. Set-up in a criss-crossed order as indicated in the picture. The arrows indicate the markings which help to place the branched tubes in the right order. Connect the fluidic unit to the pump, and place the fluidic unit in the incubator (B). Apply flow and check for correct pumping between the reservoirs. Incubate for 6 h (or the desired time).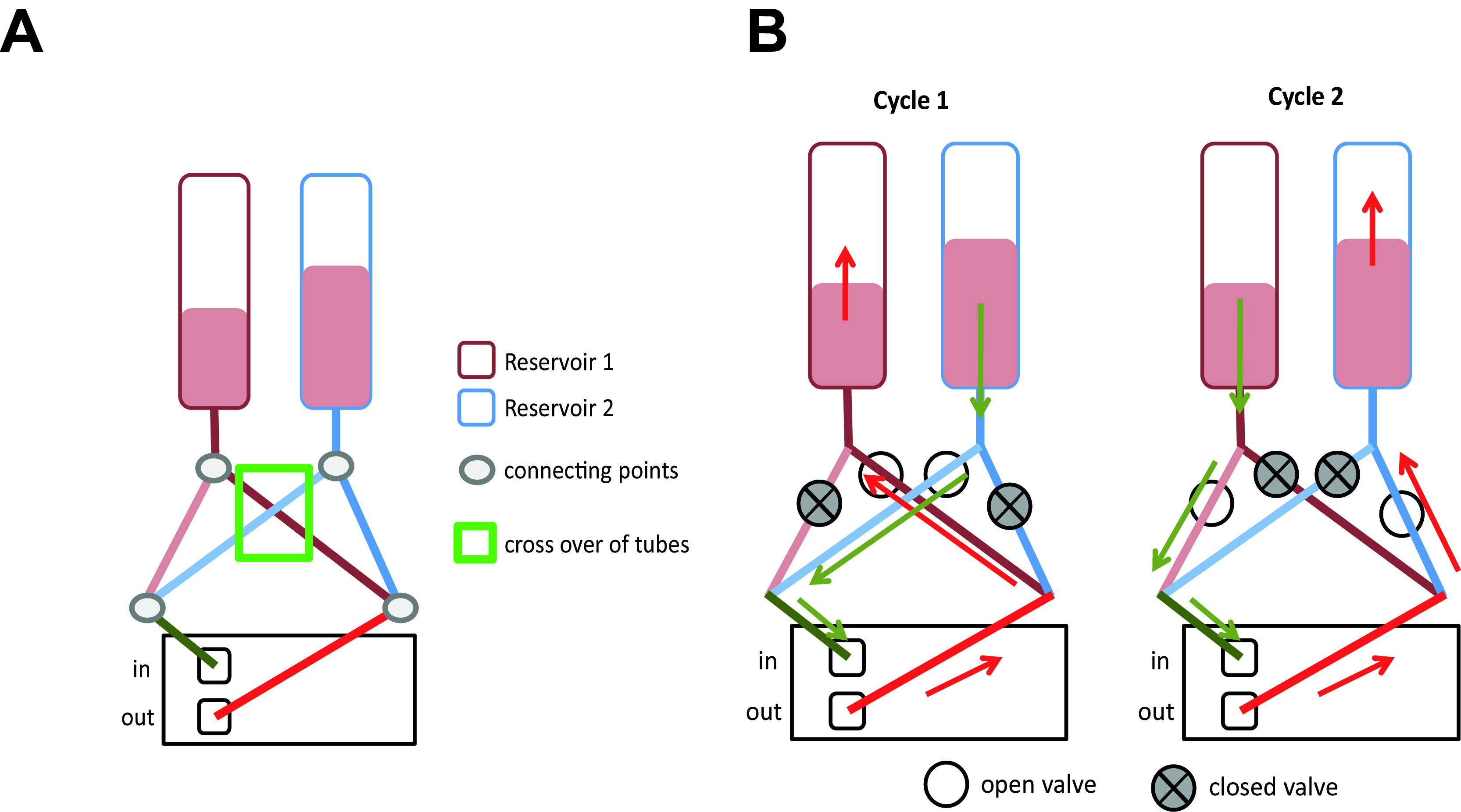
Figure 8. Principle of tubing connections and flow. A. Set-up of the tubing system. Note that from the two tubes that originate from each reservoir, one each are connected. B. Principle of the valve system and flow directions. Within the pump, the tubes from the two reservoirs that are coupled after the pump are placed in the same valve.
- Following adhesion, carefully remove the medium from the channels, wash cells once by adding 150 μl pre-warmed RPMI1640 medium and aspirating again. This will remove most non-adherent cells. Avoid vigorous pipetting as this might detach cells.
- Place the Fluidic unit including the Perfusion set and ibidi μ-slide in the incubator (Figure 7B).
- Apply flow by starting the pump via the software. Check if the flow is correct. No unbalanced medium flow between the reservoirs should occur, as this would result in aspiration of air over the time. Unbalanced medium flow can be easily detected by checking the amount of media in the two reservoirs: It leads to non-uniform medium distribution in the two reservoirs, which does not decrease but increase after starting the flow. If the flow appears to run smoothly, close the incubator door. While doing so, avoid crippling of the tubes and cables as kinks will hamper the media flow. Incubate the yeast under static/flow conditions for the desired time.
- To avoid accumulation of quorum sensing molecules and waste products in the medium of the reservoirs remove the fluidic unit from the incubator each hour during the experiment: Pause the medium flow and detach the Fluidic unit from the pump and computer. Quickly open the reservoirs under sterile conditions, remove ~10 ml (of ~11.3 ml total volume in the reservoirs) of medium. Do not aspirate too much medium to avoid air bubble formation in the tubes. Immediately replace the removed liquid with fresh RPMI1640 medium (10 ml, pre-warmed to 37 °C, equilibrated with 5% CO2).
Note: Do NOT remove the ibidi μ-slide from the fluidic unit during medium exchange! - At the end of the incubation, remove the fluidic unit from the incubator, detach the μ-slide from the tubes of the Perfusion set, and remove the medium from each channel.
- Carefully flush each channel containing cells three times each with 150 μl 1x PBS. Then, add 120 μl Histofix (or 4% paraformaldehyde) to fix the cells for 1 h at room temperature. Replace Histofix with 120 μl 1x PBS for microscopy.
Data analysis
Microscopic evaluation of filamentation was performed using a ZEISS AxioVert inverted microscope and ZEN software.
- Start the software.
- Start the microscope, place ibidi chamber on the micro slide holder and adjust.
- Make sure to choose the right magnification in the software for microscopy. You find the respective selection (for objective, zoom and Camera Adapter) in the settings ‘Microscope Components’. This is important for the software to calculate the correct μm length from pixels.
- Take pictures. Make sure to choose several random spots throughout the whole channel for quantification of filament length during microscopy. In our study (Polke et al., 2017) we analyzed the length of 70 random cells in each channel per replicate (five replicates in total).
- For analysis of filament length, set the ‘Scaling’ to ‘theoretic’ and choose the desired unit (μm is usually appropriate) in the ‘Scaling’ settings.
- To measure filament length, select the tool ‘active curve’ in the folder ‘Graphics’. This allows an exact filament length determination. This is important as hyphae are not always growing straight, but may be curvy (see Figure 9), which would falsify the filament length determination when measured in a straight line. For filament length measurement, leave out the mother yeast, but include budding yeast which have not yet separated (pseudohyphae; see Figure 9).
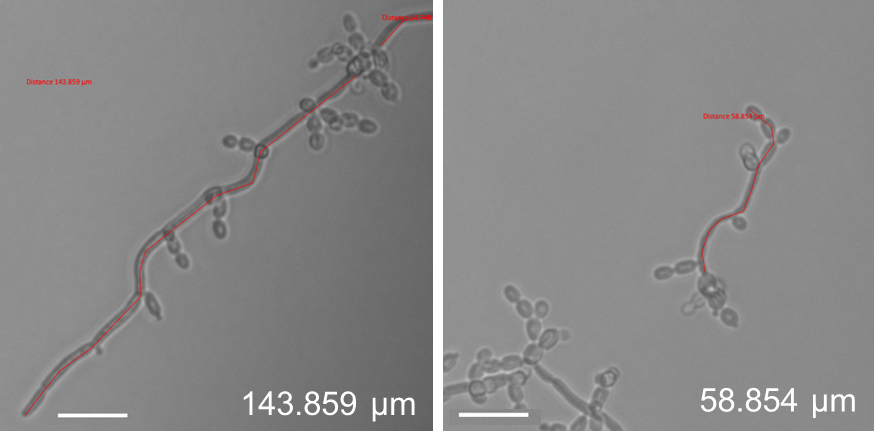
Figure 9. Filament length measurement of C. albicans eed1∆ cells grown under flow (left) or static (right) conditions using ZEN software (ZEISS). The length of each filament was measured using the active curve tool of the ZEN software (Blue edition, ZEISS). The measured part of the filaments is indicated by the red line. - Measure the length of filaments grown under static and flow conditions. Alternatively, filament induction can be determined by counting filament numbers in relation to the total cell number.
- In our study, the filament lengths from static or flow conditions were analyzed statistically by the non-parametric Mann-Whitney test using GraphPad Prism 5 software (see Figure 10).
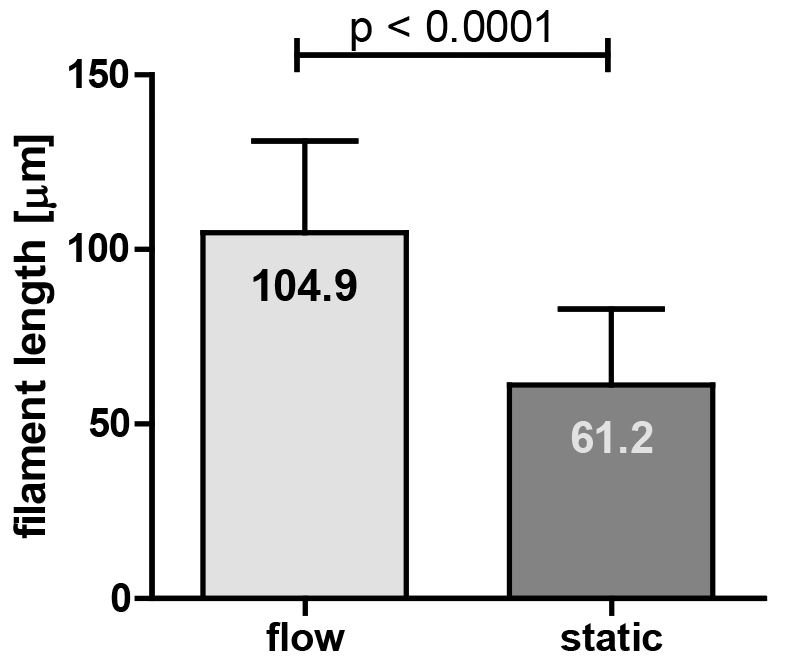
Figure 10. Quantification of the filament length of a C. albicans EED1 deletion strain (eed1∆) grown under flow or static conditions (graph taken from [Polke et al., 2017]). Filament lengths from static or flow conditions were analysed statistically by the non-parametric Mann-Whitney test using GraphPad Prism 5 software. The numbers in the bars indicate the average filament length from 70 cells in three independent biological experiments under the given growth conditions.
Notes
- Some of the ibidi® pump system materials can be sterilized by autoclaving and reused for the experimental setup. These include the tubings, and the attached reservoirs, but not the mounted filters. For sterilization, place the tubings and reservoirs in a heat stable autoclaving bag (e.g., disposal bag), close the bag using Steam Indicator Tape and sterilize in a standard laboratory autoclave at 121 °C for 20 min. For sterilization of the filters, wipe the filters several times with 70% ethanol and subsequently sterilize under UV light in a UV-crosslinker (120 mJ per cm2) or a comparable UV device for 15 min. However, we recommend not to reuse tubings and reservoirs for more than three times.
- In the flow assay system used for the experiments in our publication, only one fluidic unit (ibidi) was available and installed to generate a uni-directional flow on cells. However, one ibidi µ-slide VI0.4 has six channels for cell seeding. Thus, it would be possible to grow more than one sample under flow conditions if several fluidic units are installed. Always include a static sample as control on the slide if applicable to your scientific question.
- While we used the ibidi® pump system to generate a uni-directional flow to wash of accumulating QSMs in the cell culture supernatant, the system may also be used to simulate flow on a biofilm surface, to simulate blood flow (Wilson and Hube, 2010) and for many more applications. A detailed transcription on the principal of the ibidi® pump system, the uni-directional flow assurance and more applications using the pump system are provided on the manufacturer’s homepage (http://ibidi.com/).
- Other flow systems from other manufacturers can of course be used for this type of experiments. Important aspects when choosing a system for the experiment described here are the ease with which medium can be removed from the system, and the total medium volume in relation to the channel volume, as this determines the dilution rate. For the ibidi® pump system, for example, also bigger reservoir sets (50 ml) are available.
Recipes
- 30% glycerin solution (100 ml)
30 ml glycerol, ROTIPURAN®, water-free
Add 70 ml deionized water
Autoclave - 20% D(+)-glucose solution (500 ml)
100 g D(+)-glucose solution, water-free
Add deionized water to 500 ml
Autoclave - YPD broth (for 500 ml)
2% (w/v) (10 g) BactoTM peptone
1% (w/v) (5 g) yeast extract, micro-granulated
Add deionized water to 450 ml, autoclave
Add 50 ml sterile 20% D(+)-glucose solution (final concentration 2%) - YPD agar medium (500 ml)
2% (w/v) (10 g) BactoTM peptone
1% (w/v) (5 g) yeast extract, micro-granulated
2% (w/v) (10 g) D(+)-glucose
2% (w/v) Agar-agar, Kobe I
Autoclave - 10x PBS
1.5 M NaCl
100 mM Na2HPO4·2H2O
12 mM KH2PO4
Adjust pH to 7.4
Autoclave - 1x PBS
100 ml 10x PBS, add 900 ml sterile water - 70% ethanol (for 1 L)
Mix 700 ml ethanol (denatured, ≥ 99.8%) and 300 ml deionized water
Acknowledgments
This work has been financially supported by the Studienstiftung des deutschen Volkes e.V. (to MP) and the Deutsche Forschungsgemeinschaft (DFG JA1960/1-1 to IJ). The protocol was adapted from Wilson and Hube (2010). We thank Dr. Duncan Wilson and Dr. Sascha Brunke for helpful suggestions on the ibidi® pump system setup.
References
- Hornby, J. M., Jensen, E. C., Lisec, A. D., Tasto, J. J., Jahnke, B., Shoemaker, R., Dussault, P. and Nickerson, K. W. (2001). Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol 67(7): 2982-2992.
- Lindsay, A. K., Deveau, A., Piispanen, A. E. and Hogan, D. A. (2012). Farnesol and cyclic AMP signaling effects on the hypha-to-yeast transition in Candida albicans. Eukaryot Cell 11(10): 1219-1225.
- Polke, M., Sprenger, M., Scherlach, K., Alban-Proano, M. C., Martin, R., Hertweck, C., Hube, B. and Jacobsen, I. D. (2017). A functional link between hyphal maintenance and quorum sensing in Candida albicans. Mol Microbiol 103(4): 595-617.
- Wilson, D. and Hube, B. (2010). Hgc1 mediates dynamic Candida albicans-endothelium adhesion events during circulation. Eukaryot Cell 9(2): 278-287.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Polke, M. and Jacobsen, I. D. (2017). A Flow-assay for Farnesol Removal from Adherent Candida albicans Cultures. Bio-protocol 7(19): e2562. DOI: 10.21769/BioProtoc.2562.
Category
Microbiology > Microbial signaling > Quorum sensing
Cell Biology > Cell signaling > Quorum sensing
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










