- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of Local pH Differences within Living Salmonella Cells by High-resolution pH Imaging Based on pH-sensitive GFP Derivative, pHluorin(M153R)
Published: Vol 7, Iss 17, Sep 5, 2017 DOI: 10.21769/BioProtoc.2529 Views: 7744
Reviewed by: Dennis NürnbergEunsook ParkRon Saar-Dover

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Rapid Detection of Proliferative Bacteria by Electrical Stimulation
Conor LA Edwards [...] Munehiro Asally
Feb 5, 2020 6559 Views

Tracking the Subcellular Localization of Surface Proteins in Staphylococcus aureus by Immunofluorescence Microscopy
Salvatore J. Scaffidi [...] Wenqi Yu
May 20, 2021 7803 Views

Calibrating Fluorescence Microscopy With 3D-Speckler (3D Fluorescence Speckle Analyzer)
Chieh-Chang Lin and Aussie Suzuki
Aug 20, 2024 1950 Views
Abstract
The bacterial flagellar type III protein export apparatus is composed of a transmembrane export gate complex and a cytoplasmic ATPase complex. The export apparatus requires ATP hydrolysis and the proton motive force across the cytoplasmic membrane to unfold and transport flagellar component proteins for the construction of the bacterial flagellum (Minamino, 2014). The export apparatus is a proton/protein antiporter that couples the proton flow with protein transport through the gate complex (Minamino et al., 2011). A transmembrane export gate protein, FlhA, acts as an energy transducer along with the cytoplasmic ATPase complex (Minamino et al., 2016). To directly measure the proton flow through the FlhA channel that is coupled with the flagellar protein export, we have developed an in vivo pH imaging system with high spatial and pH resolution (Morimoto et al., 2016). Here, we describe how we measure the local pH near the export apparatus in living Salmonella cells (Morimoto et al., 2016). Our approach can be applied to a wide range of living cells. Because local pH is one of the most important parameters to monitor cellular activities of living cells, our protocol would be widely used for diverse areas of life sciences.
Keywords: Intracellular pHBackground
The proton channel activities of transmembrane proton channel complexes have been detected as reduction in cytoplasmic pH of bacterial cells (Morimoto et al., 2010; Che et al., 2014; Furukawa et al., 2017). However, to measure the proton channel activity of membrane complexes in living cells in detail, precise measurements of the local cytoplasmic pH are required. A derivative of the green fluorescence protein (GFP), pHluorin, with excitation at wavelengths of 410 and 470 nm and emission at 508 nm is a useful probe to measure the cytoplasmic pH in living cells (Miesenböck et al., 1998). This probe enables us to measure the intracellular pH precisely and quantitatively because the fluorescent intensity ratio of the two excitation wavelengths R410/470 shows a remarkable pH dependence. pHluorin can be fused genetically to a target protein to monitor the local pH around the protein. However, proteolytic cleavage often removes pHluorin from its fusion protein, not only causing a poor signal-to-noise ratio but also leading to the wrong interpretations of the data. We have found that the mutation M153R in pHluorin significantly stabilizes its fusion products while retaining the marked pH dependence of the 410/470 nm excitation ratio of the fluorescence intensity (Morimoto et al., 2011). We therefore used pHluorin(M153R) to develop a method to measure the local cytoplasmic pH in living bacterial cells in a highly quantitative and precise manner (Morimoto et al., 2016).
Materials and Reagents
- 1.5 ml microcentrifuge tubes
- Glass test tubes (IWAKI, catalog number: A-18P )
- Double-sided tape (NICHIBAN, catalog number: NW-5 )
- 24 x 32 mm coverslips (thickness: 0.12-0.17 mm) (Matsunami Glass, catalog number: C024321 )
- 18 x 18 mm coverslips (thickness: 0.12-0.17 mm) (Matsunami Glass, catalog number: C018181 )
- Pipette tips
- Filter paper (Toyo Roshi Kaisha, ADVANTEC, catalog number: 00011090 )
- Salmonella YVM1004 strain (pHluorin(M153R)-fliG) (Morimoto et al., 2011)
- Salmonella YVM1049 strain (∆fliH-fliI flhB(P28T) pHluorin(M153R)-fliG) (Morimoto et al., 2016)
- Salmonella SJW1103 strain (wild type for motility and chemotaxis) (Yamaguchi et al., 1984)
- Salmonella SJW1368 strain (∆(cheW-flhD); flagellar master operon mutant) (Ohnishi et al., 1994)
- pYVM008 (pTrc99A/pHluorin(M153R)-FliG) (Morimoto et al., 2016)
- Purified pHluorin(M153R)-FliG-His6 protein in PBS
Note: The pHluorin(M153R)-FliG-His protein is purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography from the soluble fractions of E. coli BL21(DE3) cells overexpressing pHluorin(M153R)-FliG-His as described by Minamino et al. (2000). - Ampicillin sodium salt (Wako Pure Chemical Industries, catalog number: 014-23302 )
- Gramicidin (Thermo Fisher Scientific, catalog number: G6888 )
- Potassium benzoate (Wako Pure Chemical Industries, catalog number: 164-19342 )
- Bacto tryptone (BD, BactoTM, catalog number: 211705 )
- Yeast extract (BD, BactoTM, catalog number: 212750 )
- Sodium chloride (NaCl) (Wako Pure Chemical Industries, catalog number: 192-13925 )
- Bacto agar (BD, BactoTM, catalog number: 214010 )
- Potassium dihydrogen phosphate (KH2PO4) (Wako Pure Chemical Industries, catalog number: 164-22635 )
- Dipotassium hydrogen phosphate (K2HPO4) (Wako Pure Chemical Industries, catalog number: 164-04295 )
- Ethylenediamine-N,N,N’,N’-tetraacetic acid disodium salt dihydrate (2NA) (EDTA) (Dojindo, catalog number: N001 )
- LB medium (see Recipes)
- TB medium (see Recipes)
- LB agar plate (see Recipes)
- Motility buffer (see Recipes)
Equipment
- pH meter (Beckman Coulter, model: Φ34 )
- Shaking incubator (30 °C, at 200 rpm) (TAITEC, model: BR-40LF )
- Spectrophotometer (able to measure OD600) (Shimadzu, model: UV-1800 )
- Centrifuge (able to hold 1.5 ml tubes, spin at 6,000 x g) (TOMY SEIKO, model: MX-305 )
- Single channel pipettes (1,000 µl, 100 µl) (PIPETMAN® Classic; Gilson, models: P1000 and P100 )
- Inverted fluorescence microscope (Olympus, model: IX71 ) (see Figure 1)
- 150x oil immersion objective lens (Olympus, model: UApo150XOTIRFM , NA1.45)
- Electron-multiplying charge-coupled device (EMCCD) camera (Hamamatsu Photonics, model: C9100-02 )
- Excitation filter (Omega Optical, model: 400AF30 )
- Excitation filter (Olympus, model: BP470-490 )
- Emission filter (Omega Optical, model: 520DF40 )
- High-speed wavelength switcher (Sutter Instrument, model: Lambda DG-4 )
- Dichroic mirror (Semrock, model: FF510-Di01-25x36 )
Software
- MetaMorph (Molecular Devices)
- ImageJ (National Institutes of Health, https://imagej.nih.gov/ij/)
- IGOR Pro (WaveMetrics)
- Microsoft Excel (Microsoft)
Procedure
Note: All procedures are carried out at room temperature unless otherwise specified.
- Fluorescence microscopy
- Pick a single colony of Salmonella cells expressing pHluorin(M153R)-FliG (YVM1004 and YVM1049 for local pH measurements and SJW1368 transformed with pYVM008 for in vivo calibration) from LB agar plate and inoculate it into a glass test tube with 5 ml LB and shaking at 150 rpm at 30 °C overnight. When SJW1368/pYVM008 cells are grown, ampicillin is added to a final concentration of 100 µg/ml.
- Transfer 50 µl of overnight culture to 5 ml of fresh TB and incubate at 30 °C with shaking at 150 rpm until the cell density reaches an OD600 of ca. 1.8.
Note: It takes about 15 h. - Transfer 500 µl of each culture to a 1.5 ml microcentrifuge tube and then collect the cells by centrifugation (6,000 x g, 2 min).
- Discard supernatant.
- Resuspend the cell pellet in 1.0 ml of motility buffer.
- Centrifuge at 6,000 x g for 2 min.
- Discard supernatant.
- Repeat steps A5-A7 twice.
- Resuspend the cells in 500 µl of fresh motility buffer.
- Make a tunnel slide by sandwiching double-sided tape between a 24 x 32 mm coverslip (bottom side) and an 18 x 18 mm coverslip (top side) (see Video 1).Video 1. Preparation for a tunnel slide (from Morimoto et al., 2017)
- Add 50 µl of the cell suspension to the tunnel slide and leave for 5 min to attach the cells to the coverslip surface.
- Wash out unbound cells by supplying 100 µl of the motility buffer using a pipette (P100). Absorb the excess amount of the buffer with a piece of a filter paper. (see Video 1)
- Put the sample on a custom-built inverted fluorescence microscope (Figure 1).
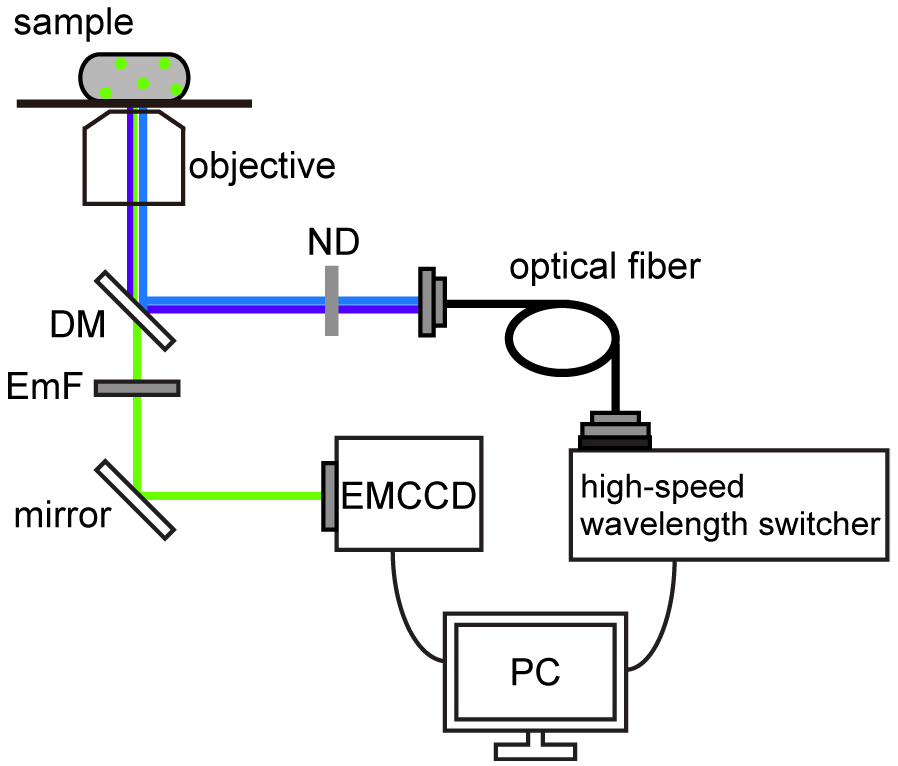
Figure 1. Schematic of pH imaging system using ratiometric pHluorin(M153R). The pH imaging system was built on an inverted fluorescence microscope with a 150x oil immersion objective lens, 1.6x variable inserts and a C9100-02 EMCCD camera. The fluorescent spots of pHluorin(M153R)-FliG were excited by a xenon lamp with two excitation filters, 400AF30 (Omega Optical) for 410-nm excitation and BP470-490 (Olympus) for 470-nm excitation. A high-speed wavelength switcher was used to switch these two excitation filters. Fluorescence emission was passed through a dichroic mirror (DM) (FF510-Di01-25x36, Semrock) and an emission filter (EmF) (520DF40, Omega Optical). Each fluorescent image was captured by the EMCCD camera. The high-speed wavelength switcher and the EMCCD camera were controlled by the MetaMorph software. ND: neutral density filter. - Select a 150x objective and insert a 1.6x variable lens.
Note: High magnification is required to measure the local cytoplasmic pH in living bacterial cells (150x objective and 1.6x variable lens with C9100-02 EMCCD [cell size: 8 x 8 µm]: 33 nm/pixel). - Switch between 410 and 470 nm excitation filters using a high-speed wavelength switcher.
- Capture each image of the pHluorin(M153R)-FliG fluorescent spots by the EMCCD camera with a 5 sec exposure time (Figure 2).
Notes:- The culture is diluted with motility buffer to adjust the number of cells to about 10 cells per 10 x 10 µm image field.
- Fluorescent spots are excited by a xenon lamp through an ND filter to avoid photobleaching. Time-lapse measurements are required to test the effect of photobleaching.
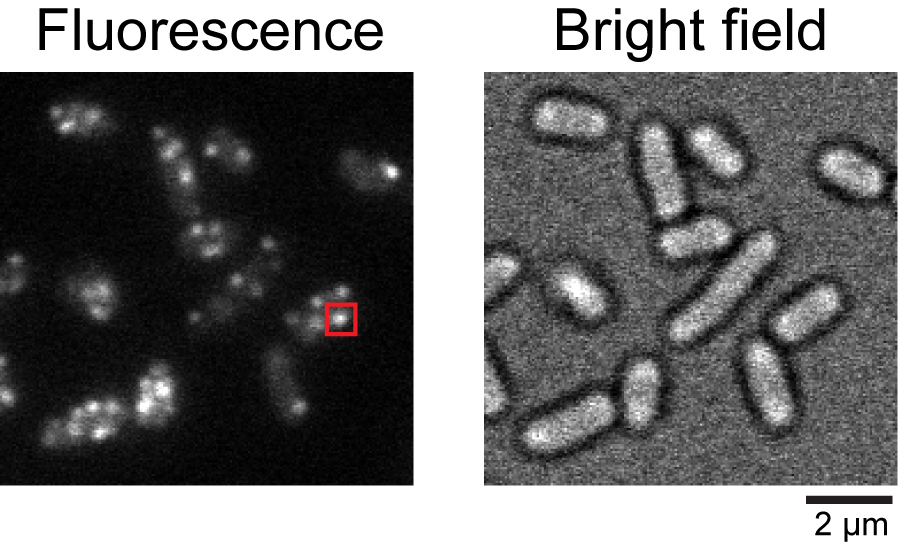
Figure 2. Typical fluorescence image of Salmonella cells expressing pHluorin(M153R)-FliG. Fluorescence and bright field images of the Salmonella YVM1004 strain. The red square indicates a typical fluorescent spot of pHluorin(M153R)-FliG.
- The culture is diluted with motility buffer to adjust the number of cells to about 10 cells per 10 x 10 µm image field.
- Pick a single colony of Salmonella cells expressing pHluorin(M153R)-FliG (YVM1004 and YVM1049 for local pH measurements and SJW1368 transformed with pYVM008 for in vivo calibration) from LB agar plate and inoculate it into a glass test tube with 5 ml LB and shaking at 150 rpm at 30 °C overnight. When SJW1368/pYVM008 cells are grown, ampicillin is added to a final concentration of 100 µg/ml.
- In vivo calibration
- After step A8: Resuspend SJW1368 cells carrying pYVM008 in 500 µl of motility buffer containing 20 µM gramicidin and 20 mM potassium benzoate.
- Add 50 µl of the cell suspension to the tunnel slide and leave for 5 min.
- Wash out unbound cells by supplying 100 µl of motility buffer containing 20 µM gramicidin and 20 mM potassium benzoate. Absorb the excess amount of the buffer using a filter paper.
- Put the sample on the fluorescence microscope.
- Acquire ratiometric fluorescence images of the cells at external pH 6.0, 6.5, 7.0 or 7.5 in the presence of 20 µM gramicidin and 20 mM potassium benzoate. The tunnel slide and the cells are freshly prepared for each external pH measurement.
- After step A8: Resuspend SJW1368 cells carrying pYVM008 in 500 µl of motility buffer containing 20 µM gramicidin and 20 mM potassium benzoate.
- In vitro calibration
- Suspend purified pHluorin(M153R)-FliG proteins in motility buffer with distinct pH values, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0 or 8.5.
- Add 50 µl of the solution containing pHluorin(M153R)-FliG proteins to the tunnel slide and absorb the excess amount of the solution with a piece of filter paper.
- Acquire fluorescence images of the pHluorin(M153R)-FliG protein solution excited by 410 and 470 nm at each pH values.
Note: When the protein concentration of pHluorin(M153R)-FliG is too high, its fluorescence intensity is considerably high, thereby saturating the sensor chip of the EMCCD camera. Therefore, it is necessary to adjust the protein concentration of pHluorin(M153R)-FliG to ca. ~100 µg/ml in order to make an in vitro calibration curve.
- Suspend purified pHluorin(M153R)-FliG proteins in motility buffer with distinct pH values, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0 or 8.5.
Data analysis
Analyze fluorescent images by image analysis software IGOR Pro or ImageJ.
- Calibration curve
- Define the total fluorescence intensity of the cell body determined by the image profile of the SJW1368 cells carrying pYVM008 (Figure 3).
Note: Since the flhDC deletion strain produces no flagella, the pHluorin(M153R)-FliG probe freely diffuses in the cytoplasm.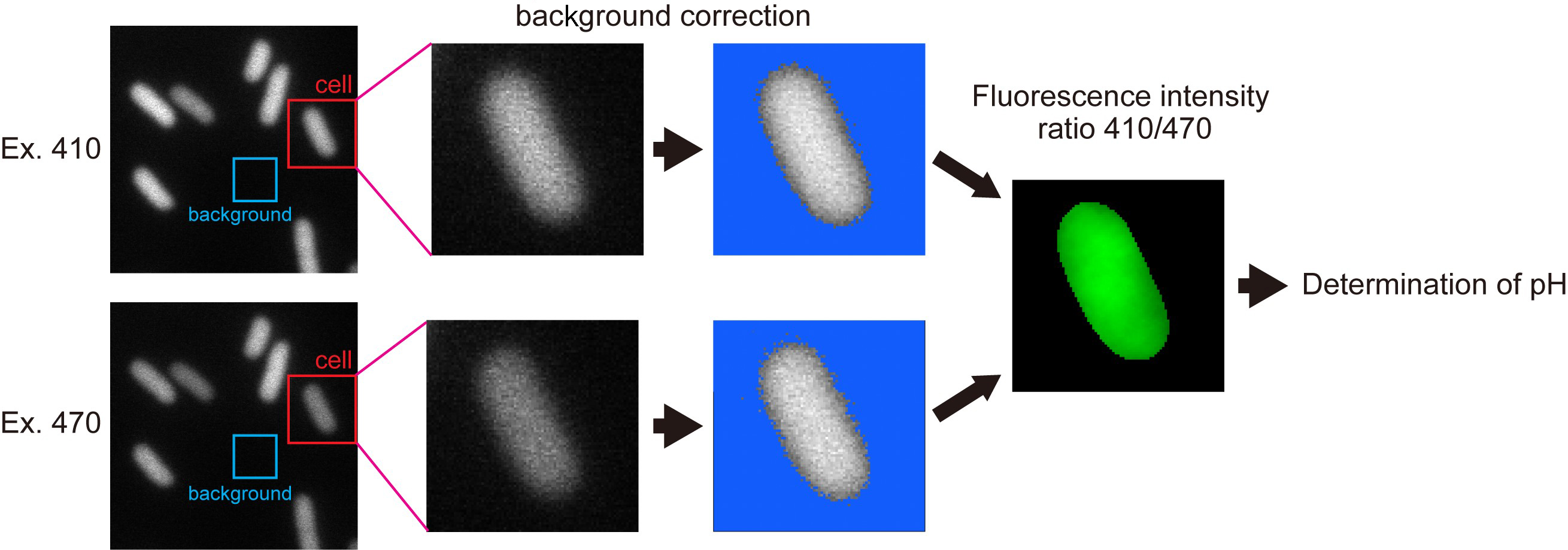
Figure 3. Typical image processes for determination of cytoplasmic bulk pH by pHluorin(M153R)-FliG. See text for details. - Define the instrumental background intensity as the mean pixel intensity within a 50 x 50 pixel ROI (region of interest) of a nearby cell-less region.
- Define the autofluorescence intensity as the mean pixel intensity of 50 wild-type SJW1103 cells producing no fluorescent proteins.
- Subtract the instrumental background and the autofluorescence of the cells from the total fluorescence intensity of the fluorescent cell body to define the fluorescence intensity of freely diffusing pHluorin(M153R)-FliG within the cells at 410 and 470 nm excitation wavelengths.
- Calculate the fluorescence intensity ratio, R410/470, of the pHluorin(M153R)-FliG probe.
- Determine the cytoplasmic pH from the R410/470 of the pHluorin(M153R)-FliG that was obtained at each external pH in the presence of 20 µM gramicidin and 20 mM potassium benzoate, both of which equilibrate the pH inside and outside Salmonella cells, to make the in vivo calibration curve (Figure 4).
Note: In vivo calibration clearly shows that pHluorin(M153R)-FliG works as a pH indicator in living cells.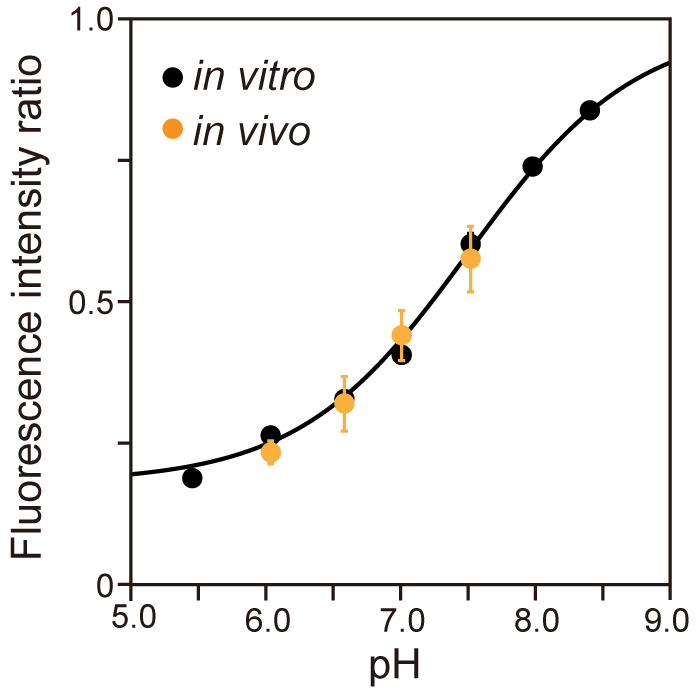
Figure 4. Calibration curve of pHluorin(M153R)-FliG in vivo and in vitro. Fluorescent images of purified pHluorin(M153R)-His protein solution were taken by the pH imaging system over a pH range from 5.5 to 8.5. The fluorescent intensity ratio 410/470 was calculated at each pH value (black, in vitro), and the data were fitted by a sigmoid function. Then, Salmonella SJW1368 cells harboring pYVM008 were observed by the same system in motility buffer at four distinct pH values, 6.0, 6.5, 7.0 and 7.5, in the presence of 20 µM gramicidin and 20 mM potassium benzoate, and the intracellular pH was measured (orange, in vivo). Vertical bars indicate standard deviations (Modified from Morimoto et al., 2016). - Define the fluorescence intensity of the purified pHluorin(M153R)-FliG protein after subtraction of the instrumental background intensity as the mean pixel intensity within a 100 x 100 pixel ROI of an image of the solution without fluorescent proteins.
- Calculate R410/470 of the purified pHluorin(M153R)-FliG at each pH value.
- Plot the emission intensity ratios as a function of pH to make the in vitro calibration curve over a pH range from 5.5 to 8.5 (Figure 4).
- Fit the calibration data by a sigmoid curve function using the IGOR Pro software.
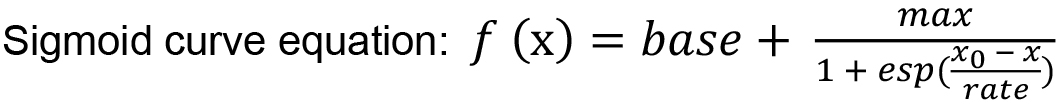
- Define the total fluorescence intensity of the cell body determined by the image profile of the SJW1368 cells carrying pYVM008 (Figure 3).
- Determination of local pH values
- Apply a rectangular mask for the fluorescent spots of the pHluorin(M153R)-FliG of 20 x 20 pixels to the ROI in a fluorescent image of the YVM1004 cells (Figure 2).
- Subtract the total background intensity including the instrumental background and the autofluorescence of the cell from each pixel value. The instrumental background intensity is defined as the mean pixel intensity within a 50 x 50 pixel ROI of a nearby cell-less region. The autofluorescence intensity of the cell is defined as the mean pixel intensity within the ROI of a non-fluorescent cell SJW1103.
- Define the fluorescence intensity of a single fluorescent spot of the pHluorin(M153R)-FliG probe as an integral fluorescent intensity value within the ROI.
- Calculate the ratio of the fluorescent intensity of the pHluorin(M153R)-FliG spots at 410 and 470 nm excitation wavelengths.
- Determine the local pH from the R410/470 value of each fluorescent spot using the in vitro calibration curve.
- Calculate the average of the local pH values and the standard error of the mean from the data of more than 100 fluorescent spots using Microsoft Excel (Microsoft) (Figure 5).
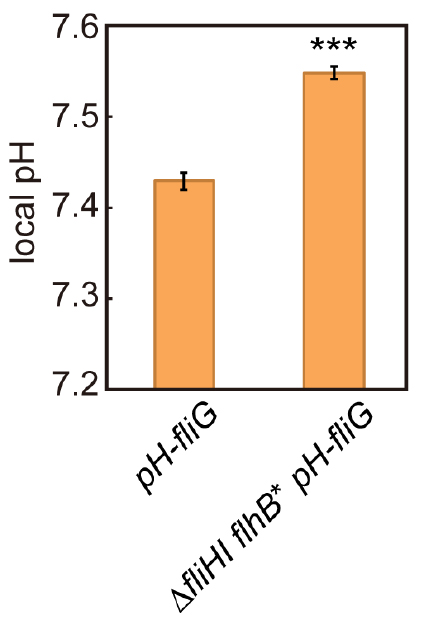
Figure 5. Measurement of local pH using pHluorin(M153R)-FliG. The local pH around the flagellar type III protein export apparatus of the pH-fliG (YVM1004) and the ∆fliHI flhB(P28T) pH-fliG (YVM1049) cells was measured. More than 300 fluorescent spots were analyzed. Vertical bars indicate standard errors (Modified from Morimoto et al., 2016).
- Apply a rectangular mask for the fluorescent spots of the pHluorin(M153R)-FliG of 20 x 20 pixels to the ROI in a fluorescent image of the YVM1004 cells (Figure 2).
Notes
Salmonella cells expressing pHluorin(M153R)-FliG should be incubated with shaking for more than 5 h to be fully matured the fluorophores of pHluorin(M153R), because sufficient fluorescence intensity is required to detect the local pH.
Recipes
- LB medium (per liter)
10 g Bacto tryptone
5 g yeast extract
5 g NaCl - TB medium (per liter)
10 g Bacto tryptone
10 mM potassium phosphate, pH 7.0 - LB agar plate
LB medium
1.5% Bacto agar - Motility buffer
10 mM potassium phosphate
0.1 mM EDTA
Adjust to different pH values: 5.5, 6.0, 6.5, 7.0, 7.5, 8.0 or 8.5
Acknowledgments
This protocol was modified from a previous work (Morimoto et al., 2016). This research was supported in part by JSPS KAKENHI Grant Numbers JP15K14498 and JP15H05593 to YVM, JP25000013 to KN and JP26293097 to TM, and MEXT KAKENHI Grant Numbers JP26115720 and JP15H01335 to YVM and JP24117004, JP25121718 and JP15H01640 to TM.
References
- Che, Y. S., Nakamura, S.,Morimoto, Y. V., Kami-ike, N., Namba, K. and Minamino, T. (2014). Load-sensitive coupling of proton translocationand torque generation in the bacterial flagellar motor. Mol Microbiol 91(1): 175-184.
- Furukawa,A., Yoshikaie, K., Mori, T., Mori, H., Morimoto, Y. V., Sugano, Y., Iwaki, S.,Minamino, T., Sugita, Y., Tanaka, Y. and Tsukazaki, T. (2017). Tunnel formation inferred from the I-form structures of the proton-drivenprotein secretion motor SecDF. Cell Rep 19(5): 895-901.
- Miesenböck, G., De Angelis, D. A. andRothman, J. E. (1998). Visualizing secretion and synaptic transmissionwith pH-sensitive green fluorescent proteins. Nature 394(6689):192-195.
- Minamino,T. (2014). Protein export through thebacterial flagellar type III export pathway. Biochim Biophys Acta 1843(8): 1642-1648.
- Minamino,T., Morimoto, Y. V., Hara, N., Aldridge, P. D. and Namba, K. (2016). The bacterial flagellar type III export gatecomplex is a dual fuel engine that can use both H+ and Na+ for flagellar protein export. PLoSPathog 12(3): e1005495.
- Minamino,T., Morimoto, Y. V., Hara, N. and Namba, K. (2011). An energy transduction mechanism used inbacterial flagellar type III protein export. Nat Commun 2: 475.
- Minamino,T., Yamaguchi, S. and Macnab, R. M. (2000). Interaction between FliE and FlgB, a proximalrod component of the flagellar basal body of Salmonella. JBacteriol 182(11): 3029-3036.
- Morimoto,Y. V., Che, Y. S., Minamino, T. and Namba, K. (2010). Proton-conductivity assay of plugged andunplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS Lett 584(6):1268-1272.
- Morimoto,Y. V., Kami-ike, N., Miyata, T., Kawamoto, A., Kato, T., Namba, K. andMinamino, T. (2016). High-resolution pH imaging of living bacterialcells to detect local pH differences. mBio 7(6).
- Morimoto,Y. V., Kojima, S., Namba, K. and Minamino, T. (2011). M153R mutation in a pH-sensitive greenfluorescent protein stabilizes its fusion proteins. PLoS One 6(5):e19598.
- Morimoto, Y. V., Namba, K. and Minamino, T.(2017). Bacterial intracellularsodium ion measurement using CoroNa Green. Bio Protoc 7(1) e2092.
- Ohnishi,K., Ohto, Y., Aizawa, S., Macnab, R. M. and Iino, T. (1994). FlgD is a scaffolding protein needed forflagellar hook assembly in Salmonella typhimurium. J Bacteriol 176(8):2272-2281.
- Yamaguchi,S., Fujita, H., Sugata, K., Taira, T. and Iino, T. (1984).Genetic analysis of H2, thestructural gene for phase-2 flagellin in Salmonella. J Gen Microbiol 130(2): 255-265.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Morimoto, Y. V., Kami-ike, N., Namba, K. and Minamino, T. (2017). Determination of Local pH Differences within Living Salmonella Cells by High-resolution pH Imaging Based on pH-sensitive GFP Derivative, pHluorin(M153R). Bio-protocol 7(17): e2529. DOI: 10.21769/BioProtoc.2529.
Category
Microbiology > Microbial cell biology > Cell imaging
Cell Biology > Cell imaging > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










