- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro AMPylation Assays Using Purified, Recombinant Proteins
Published: Vol 7, Iss 14, Jul 20, 2017 DOI: 10.21769/BioProtoc.2416 Views: 8163
Reviewed by: Peichuan ZhangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Characterization of Protein Domain Function via in vitro DNA Shuffling
Kathy Hiu Laam Po [...] Sheng Chen
Jun 5, 2018 6636 Views

A Method for SUMO Modification of Proteins in vitro
Christine C. Lee [...] Michael J. Matunis
Oct 5, 2018 8113 Views

Protocol for Spontaneous and Chaperonin-assisted in vitro Refolding of a Slow-folding Mutant of GFP, sGFP
Anwar Sadat [...] Koyeli Mapa
Jul 20, 2021 3457 Views
Abstract
Post-translational protein modifications (PTMs) orchestrate the activity of individual proteins and ensure their proper function. While modifications such as phosphorylation or glycosylation are well understood, more unusual modifications, including nitrosylation or AMPylation remain comparatively poorly characterized. Research on protein AMPylation–which refers to the covalent addition of an AMP moiety to the side chains of serine, threonine or tyrosine–has undergone a renaissance (Yarbrough et al., 2009; Engel et al., 2012; Ham et al., 2014; Woolery et al., 2014; Preissler et al., 2015; Sanyal et al., 2015; Truttmann et al., 2016; Truttmann et al., 2017). The identification and characterization of filamentation (fic) domain-containing AMPylases sparked new interest in this PTM (Kinch et al., 2009; Yarbrough et al., 2009). Based on recent in vivo and in vitro studies, we now know that secreted bacterial AMPylases covalently attach AMP to members of the Rho family of GTPases, while metazoan AMPylases modify HSP70 family proteins in the cytoplasm and the endoplasmic reticulum (ER) (Itzen et al., 2011; Hedberg and Itzen, 2015; Truttmann and Ploegh, 2017). AMPylation is thought to trap HSP70 in a primed yet transiently disabled state that cannot participate in protein refolding reactions (Preissler et al., 2015). In vitro AMPylation experiments are key to assess the activity, kinetics and specificity of protein AMPylation catalyzed by pro- and eukaryotic enzymes. These simple assays require recombinant AMPylases, target proteins (Rho GTPases, HSP70s), as well as ATP as a nucleotide source. Here, we describe strategies to qualitatively and quantitatively study protein AMPylation in vitro.
Keywords: AMPylationBackground
Metazoan cell signaling is complex and requires tight control. Aberrations in this well-balanced system threaten cellular homeostasis and induce several maintenance systems aimed at restoring the balance (Kim et al., 2013). Protein AMPylation is directly linked to cellular stress: AMPylation of Rho GTPases by bacterial toxins rewires GTPase-dependent signaling, eventually leading to a collapse of the actin cytoskeleton and cell death (Yarbrough et al., 2009; Mattoo et al., 2011). In contrast, AMPylation of Grp78/BiP in the ER keeps this chaperone in a primed, yet silent conformation to be awoken and set in motion once the burden of unfolded protein in the ER surpasses a certain threshold (Preissler et al., 2015; Sanyal et al., 2015). We and others have extensively used in vitro AMPylation assays to study general properties, target selectivity as well as reaction kinetics of Fic domain-containing AMPylases. We used a combination of distinct in vitro AMPylation assays to i) identify novel targets in complex cell lysates, ii) validate suspected targets and iii) approach the role of AMPylase dimerization and auto-modification as prerequisites for enzymatic activity (Truttmann et al., 2015; 2016 and 2017). Our efforts aim at understanding the scope and impact of the AMPylome on cellular signaling. The in vitro AMPylation assays described herein present methods to achieve this goal.
Materials and Reagents
- 1.5 ml tubes (1.5 ml Snaplock Microcentrifuge Tube) (Corning, Axygen®, catalog number: MCT-150-C-S )
- Pipette tips (Thermo Fisher Scientific, Thermo ScientificTM, catalog numbers: 9400327 , 9401255 , 9401410 )
- Microcentrifuge Tube Locks (Sorenson BioScience, catalog number: 11870 )
- Autoradiography film (Carestream Health X-OmatTM LS Film) (Eastman Kodak, catalog number: 05-728-45 )
- Saran wrap (generic)
- Whatman 3MM filter paper (GE Healthcare, catalog number: 3030-6185 )
- Sterile, deionized water (generic)
- Ice in isolated containment (generic)
- Ethanol (Sigma-Aldrich, catalog number: 362808 )
- Purified recombinant AMPylase at 1.0 μg/μl or higher (HIS6-HYPEaa187-437; homemade; see Truttmann et al., 2015)
- Purified recombinant target proteins at 1.0 μg/μl or higher (i.e., HIS6-Histone H3; homemade; see Truttmann et al., 2015)
- Appropriate TRIS-glycine gels (CriterionTM TGXTM Precast Midi Protein Gel) (Bio-Rad Laboratories, catalog number: 5671023 )
- Molecular weight marker (Precision PlusTM Protein Dual Color Standard) (Bio-Rad Laboratories, catalog number: 1610374 )
- 2-amino-2-(hydroxymethyl)-1,3-propanediol (Tris-base) (Sigma-Aldrich, catalog number: 252859 )
- 11.8 M hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 258148 )
- DL-dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43815 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- [Alpha-33P]ATP, 10 mCi/ml; 3,000 Ci/mmol (Hartman Analytic, catalog number: SRF-207 )
Note: It is of uttermost importance to use [Alpha-33P]ATP and not [Gamma-33P]ATP, which is used to study kinase-dependent phosphorylation events. - Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014 )
- Potassium chloride (KCl) (EMD Millipore, catalog number: PX1405 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: 74255 )
- 2-mercaptoethanol (Sigma-Aldrich, catalog number: M6250 )
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126 )
- Poly-Phenyl-Oxazole (PPO) (Sigma-Aldrich, catalog number: 216984 )
- Dimethyl sulfoxide (DMSO) Sigma-Aldrich, catalog number: 276855 )
- 1 M Tris-HCl (pH 7.5) (see Recipes)
- 1 M DTT (see Recipes)
- 1 M MgCl2 (see Recipes)
- 100 mM ATP (see Recipes)
- 5 M NaCl (see Recipes)
- Protein storage buffer (see Recipes)
- Reaction buffer (see Recipes)
- SDS-PAGE 6x sample buffer (see Recipes)
- DMSO/PPO solution (see Recipes)
Equipment
- Pipettes (Thermo Fisher Scientific, Thermo ScientificTM, catalog numbers: 4600170 , 4600240 and 4600250 )
- -20 °C freezer (generic)
- 4 °C refrigerator (generic)
- Geiger-counter (generic)
- Refrigerated tabletop centrifuge for 1.5 ml Eppendorf tubes (Eppendorf, model: 5810 R )
- 10 μl Hamilton syringe (Hamilton, catalog number: 80075 )
- Radiation safety gear and personal protection equipment (generic)
- Vacuum gel dryer (Bio-Rad Laboratories, model: Model 583 )
- Glass tray (generic; 5 x 10 inches at least)
- Timer (Alarm Timer) (Grainger, catalog number: 8RLR2 )
- pH meter (Thermo Fisher Scientific, Thermo ScientificTM, model: Orion StarTM A111 )
- Balance (Sartorius, model: Cubis® Precision Balance )
- Vortex (Vortex-Genie 2 Vortexer) (VWR, catalog number: VWR-VG3 )
- SDS-PAGE system (Bio-Rad Laboratories, model: CriterionTM Cell and PowerPacTM Basic Power Supply, catalog number: 1656019 )
- Autoradiography cassettes (FisherBiotech Electrophoresis Systems Autoradiography Cassette, 8 x 10 in) (Fisher Scientific, model: FBAC 810)
Note: This product is not available anymore.
Software
- Fiji/ImageJ image analysis software (https://fiji.sc/)
Procedure
Note: 33P-ATP is radioactive. Please counsel with your radio safety officer regarding your institute’s radio waste disposal and radio protection routine and ensure to obtained all required protective equipment–that should include but may not be limited to a lab coat, double-gloves and safety goggles–before starting the experiment. Continuously monitor work surfaces using a Geiger-counter and decontaminate if required following your institute’s decontamination protocol.
- In vitro AMPylation reaction
- Mix 5 μg of recombinant AMPylase enzyme in 10 μl protein storage buffer (see Recipes) with 20 μl reaction buffer (see Recipes) supplemented with 0.5 μl/sample 33P-ATP.
Notes: - The amount of AMPylase to use in an in vitro reaction depends on the relative activity of the individual enzymes; very potent enzymes (i.e., VopS, IbpA) will in vitro modify a molar excess of target protein (i.e., Rac1, Cdc42); thus, 1 μg/reaction is sufficient. In contrast, wild-type versions of metazoan AMPylases in general perform poorly in vitro; thus, 5-10 μg/reaction is preferred.
- Wear proper protective equipment and monitor work surfaces during this step with a Geiger-counter.
- Incubate reaction for 60 min at 20 °C.
Notes: - This step pre-loads or primes the enzyme, which will maximize target AMPylation.
- Throughout the protocol, suggested 20 °C incubation steps can also be performed at room temperature (approximately 20 °C).
- Centrifuge sample for 30 sec at 7,000 x g.
- Add 2 μg of target protein in 10 μl protein storage buffer to the primed enzyme (final reaction volume: 40 μl).
- Incubate reaction for 60 min at appropriate temperature (VopS, IbpA: 20 °C; FIC-1: 30 °C; HYPE: 37 °C).
Note: Reactions can be performed at 20 °C; however, incubation at higher temperatures (37 °C) enhances target AMPylation. - Quench the reaction by adding 8 μl of 6x SDS-PAGE loading buffer (see Recipes).
- Properly close 1.5 ml tubes and put on tube locks to prevent lids from opening inadvertently during step A9.
- Boil samples for 10 min at 95 °C.
Note: Tube locks are essential, as they will minimize the chance for tubes to pop open while boiling (see next step); should accidental opening of a tube have occurred, identify likely [33P]-contamination using a Geiger-counter and decontaminate according to your radiation safety office’s guidelines. - Proceed with gel electrophoresis or store samples at -80 °C.
- Mix 5 μg of recombinant AMPylase enzyme in 10 μl protein storage buffer (see Recipes) with 20 μl reaction buffer (see Recipes) supplemented with 0.5 μl/sample 33P-ATP.
- Sample analysis by gel electrophoresis and autoradiography
- Equilibrate samples on ice for 10 min.
- In the meantime, choose appropriate gel: if available, a TRIS-glycine gradient gel (i.e., 4-16%) should be used; alternatively, select the polyacrylamide percentage according to your target protein’s mass: i.e., 12% for HSP70 (approximately 70 kDa), 15% for Histones (approximately 15 kDa) (acrylamide: Bis-acrylamide = 30:0.8); prepare gel running buffer according to gel manufacturer’s instructions.
- Centrifuge samples for 30 sec at 7,000 x g.
- Load gel: add molecular weight marker and individual samples to each well of the SDS-PAGE gel.
Note: The use of gel loading tips is recommended to minimize cross-well spilling. Alternatively, use a 10 μl Hamilton syringe for loading. Load only ~50% of your total sample; store remaining half in case you need to re-run the gel. - Run the gel at 60-100 V during the stacking stage, then adjust to120 V (constant voltage) for ~2 h.
Note: The running time depends on the gel system used.Note: Steps B6-B17 are referred to as fluorography, a method developed by Bonner and Laskey (Bonner and Laskey, 1974) to be able to detect 3H in autoradiograms of SDS-PAGE gels. The method also improves sensitivity for other soft emitters, less so for higher energy isotopes such as 33P. An added advantage is that the procedure confers mechanical stability to low-percentage gels, owing to the precipitation of PPO. Gels with acrylamide percentages lower than 5% should not be subjected to fluorography, as the mechanical properties of these gels can yield an irregular final product. Commercially available solutions for impregnation of gels with fluorophores are available, often at higher cost than home-made DMSO-PPO solutions. Wear gloves at all times, since the DMSO-PPO solution can penetrate the skin and cause local precipitation of PPO in tissues.
- Carefully transfer the gel into a metal or glass tray (a baking dish will serve the purpose).
- Add 500 ml of DMSO for a large gel of 100 ml volume, smaller gels can be handled with less.
Notes:- Add ~5 times the volume of the gel of DMSO to cover the gel completely.
- Incubate at room temperature for 60 min with agitation.
- Add ~5 times the volume of the gel of DMSO to cover the gel completely.
- Discard DMSO
Note: This DMSO must be considered as potentially radioactive; dispose accordingly. - Add a fresh volume (see step B7) of DMSO and incubate for 60 min at room temperature.
- Discard DMSO.
Note: This DMSO must be considered as potentially radioactive; dispose accordingly. - Cover the gel in DMSO-PPO solution (22.2 g of PPO and 80 ml of DMSO; scale volume according to need) (see Recipes).
- Incubate for 2 h at room temperature with agitation.
- Remove DMSO-PPO as completely as possible.
Note: The DMSO-PPO solution can be recycled and reused for up to 3 gels/experiments. Replenish PPO in the DMSO-PPO solution after each cycle with an amount of PPO approximately equal to that now precipitated in the gel. - Cover the gel in the glass tray with 2 cm of ddH2O and incubate for 60 min at room temperature (Figure 1A and Video 1).
Notes:- Adding ddH2O to the dehydrated, PPO-soaked gel will result in immediate precipitation of PPO and will rapidly turn the transparent gel into a milky-white gel.
- Use a paper towel to remove any precipitated PPO from the tray.
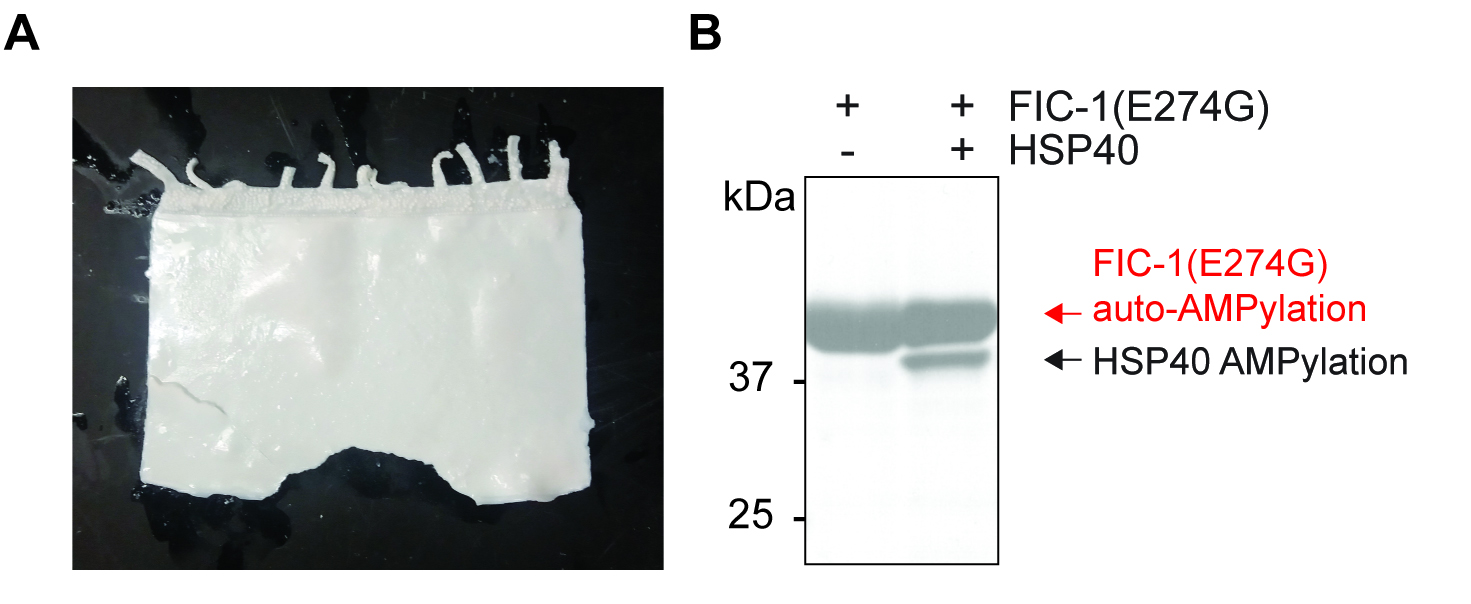
Figure 1. PPO treatment and final autoradiography plot. A. Typical appearance of SDS-PAGE gel after successful dehydration and PPO loading (see Video 1 for more details); B. Representative autoradiography plot depicting auto-AMPylation of the conferring enzyme (FIC-1(E274G), a constitutive-active FIC-1 version), as well as target modification (HSP40).
Video 1. PPO precipitation in dehydrated SDS-PAGE gel. Dehydrated and PPO-loaded SDS-PAGE gel is kept in a glass tray and distilled water is added. Upon contact with water, PPO precipitates within the gel. Excess PPO still present in the glass tray precipitates, too. To achieve best results, incubate SDS-PAGE gel for 60 min at room temperature (step B14) before proceeding with washing (step B15).
- Adding ddH2O to the dehydrated, PPO-soaked gel will result in immediate precipitation of PPO and will rapidly turn the transparent gel into a milky-white gel.
- Discard ddH2O.
- Add fresh ddH2O.
- Incubate gel in ddH2O for 10 min.
- Discard ddH2O.
- Repeat previous step 5 times.
- Pre-soak filter paper (Whatman 3MM) with ddH2O.
- Transfer gel face-up onto soaked filter paper.
- Transfer filter-gel stack to gel drying unit.
- Cover the filter-gel stack with Saran wrap.
Note: Adding a single layer of Saran wrap or similar prevents the gel from sticking the gel dryer’s rubber cover and facilitates the removal of the gel once dried onto the filter. - Dry gel for 90 min at 65 °C. It is essential that a strong vacuum be maintained to avoid cracking of the gel. If necessary, use a lyophilizer-style set-up with a cold trap.
- Transfer gel into autoradiography cassette.
Note: At this stage, it is sometimes worthwhile to use a Geiger-counter to get a feeling for the signal intensity attained; if audible with a sensitive Geiger-counter, an overnight exposure of the film will usually yield an interpretable signal. - In a dark room, expose to film.
- Expose autoradiogram for 10 min to 10 days, depending on the expected signal intensity.
Note: This is a rather arbitrary factor and often must be evaluated empirically; 33P has a half-life time of ~30 days. - If the film is exposed for longer than 12 h, store the autoradiography cassette at -80 °C to enhance sensitivity of detection.
- Develop film according to manufacturer’s instructions (Figure 1B).
- Equilibrate samples on ice for 10 min.
Data analysis
- In vitro AMPylation assays as presented in this protocol will primarily result in a qualitative assessment of target AMPylation. It is strongly recommended to repeat experiments and to verify target AMPylation in three independent replica.
- If required, protein AMPylation can be quantified as follows:
- Scan develop autoradiography film.
- Open scan image in Fiji/ImageJ image analysis software (https://fiji.sc/).
- Use the integrated analyze ==> gels plugin and redeem the intensities of individual bands depicting AMPylated proteins.
- Determine average and standard deviation combining data from at least 3 independent replica.
- Use appropriate statistical methods to evaluate significance of observed AMPylation levels (e.g., Student’s t-test).
Notes: - The appropriate statistical test will depend on the study design
- Users are strongly encouraged to consult available online tutorials prior to using this tool for the first time (https://www.youtube.com/watch?v=JlR5v-DsTds).
Notes
- If performed under identical experimental conditions, in vitro AMPylation assays are highly reproducible and should provide a similar qualitative assessment of target modification.
- [33P] has a half-life time of 25.3 days; thus, signal intensity of particles emitted from AMPylated targets will decrease with daily increments. If quantification of signal intensities is desired, it might be necessary to use relative (intensity as compared to an internal control) rather than absolute quantification strategies.
Recipes
- 1 M Tris-HCl (pH 7.5)
For 100 ml:
12.1 g Tris-base
90 ml ddH2O
Adjust to pH 7.5 with HCl
Adjust volume with ddH2O to 100 ml
Store at room temperature - 1 M DTT
For 100 ml:
15 g DTT in 100 ml ddH2O
Store in aliquots at -20 °C - 1 M MgCl2
For 100 ml:
47.6 g MgCl2
Adjust volume with ddH2O to 100 ml
Store in aliquots at -20 °C - 100 mM ATP
For 100 ml:
5.51 g ATP
Adjust volume with ddH2O to 100 ml
Store in aliquots at -20 °C - 5 M NaCl
For 100 ml:
29.22g NaCl in 100 ml ddH2O - Protein storage buffer (50 mM Tris-HCl pH7.5, 150 mM NaCl, 10% glycerol [v/v])
For 10 ml:
500 μl 1 M Tris-HCl pH 7.5
300 μl 5 M NaCl
1 ml glycerol
Adjust volume with ddH2O to 10 ml - Reaction buffer (50 mM TRIS-HCl pH 7.5, 10 mM MgCl2, 150 mM NaCl, 2 mM DTT)
For 10 ml:
500 μl 1 M Tris-HCl pH 7.5
100 μl 1 M MgCl2
300 μl 5 M NaCl
20 μl 1 M DTT
9.08 ml ddH2O - SDS-PAGE 6x sample buffer (375 mM Tris-HCl pH 6.8, 6% SDS [w/v], 48% glycerol [v/v], 9% 2-mercaptoethanol [v/v], 0.03% bromophenol blue [w/v])
For 100 ml:
5.91 g Tris-HCl
6 g SDS
48 ml 100% glycerol
9 ml 14.7 M 2-mercaptoethanol
30 mg bromophenol blue - DMSO/PPO solution
30 g PPO in 120 ml DMSO
Acknowledgments
The authors like to thank the members of the Ploegh lab for helpful discussions and input. This work was supported by awards from the National Institute of Health to H.L.P. M.C.T. is supported by a Young Investigator Award from Emerald Foundation, Inc. This protocol was modified from previous work as described in Truttmann et al., 2016.
References
- Bonner, W. M. and Laskey, R. A. (1974). A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem 46(1): 83-88.
- Engel, P., Goepfert, A., Stanger, F. V., Harms, A., Schmidt, A., Schirmer, T. and Dehio, C. (2012). Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature 482(7383): 107-110.
- Ham, H., Woolery, A. R., Tracy, C., Stenesen, D., Kramer, H. and Orth, K. (2014). Unfolded protein response-regulated Drosophila Fic (dFic) protein reversibly AMPylates BiP chaperone during endoplasmic reticulum homeostasis. J Biol Chem 289(52): 36059-36069.
- Hedberg, C. and Itzen, A. (2015). Molecular perspectives on protein adenylylation. ACS Chem Biol 10(1): 12-21.
- Itzen, A., Blankenfeldt, W. and Goody, R. S. (2011). Adenylylation: renaissance of a forgotten post-translational modification. Trends Biochem Sci 36(4): 221-228.
- Kim, Y. E., Hipp, M. S., Bracher, A., Hayer-Hartl, M. and Hartl, F. U. (2013). Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 82: 323-355.
- Kinch, L. N., Yarbrough, M. L., Orth, K. and Grishin, N. V. (2009). Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS One 4(6): e5818.
- Mattoo, S., Durrant, E., Chen, M. J., Xiao, J., Lazar, C. S., Manning, G., Dixon, J. E. and Worby, C. A. (2011). Comparative analysis of Histophilus somni immunoglobulin-binding protein A (IbpA) with other fic domain-containing enzymes reveals differences in substrate and nucleotide specificities. J Biol Chem 286(37): 32834-32842.
- Preissler, S., Rato, C., Chen, R., Antrobus, R., Ding, S., Fearnley, I. M. and Ron, D. (2015). AMPylation matches BiP activity to client protein load in the endoplasmic reticulum. Elife 4: e12621.
- Sanyal, A., Chen, A. J., Nakayasu, E. S., Lazar, C. S., Zbornik, E. A., Worby, C. A., Koller, A. and Mattoo, S. (2015). A novel link between Fic (filamentation induced by cAMP)-mediated adenylylation/AMPylation and the unfolded protein response. J Biol Chem 290(13): 8482-8499.
- Truttmann, M. C., Cruz, V. E., Guo, X., Engert, C., Schwartz, T. U. and Ploegh, H. L. (2016). The Caenorhabditis elegans protein FIC-1 is an AMPylase that covalently modifies heat-shock 70 family proteins, translation elongation factors and histones. PLoS Genet 12(5): e1006023.
- Truttmann, M. C. and Ploegh, H. L. (2017). rAMPing up stress signaling: Protein AMPylation in metazoans. Trends Cell Biol.
- Truttmann, M. C., Wu, Q., Stiegeler, S., Duarte, J. N., Ingram, J. and Ploegh, H. L. (2015). HypE-specific nanobodies as tools to modulate HypE-mediated target AMPylation. J Biol Chem 290(14): 9087-9100.
- Truttmann, M. C., Zheng, X., Hanke, L., Damon, J. R., Grootveld, M., Krakowiak, J., Pincus, D. and Ploegh, H. L. (2017). Unrestrained AMPylation targets cytosolic chaperones and activates the heat shock response. Proc Natl Acad Sci U S A 114(2): E152-E160.
- Woolery, A. R., Yu, X., LaBaer, J. and Orth, K. (2014). AMPylation of Rho GTPases subverts multiple host signaling processes. J Biol Chem 289(47): 32977-32988.
- Yarbrough, M. L., Li, Y., Kinch, L. N., Grishin, N. V., Ball, H. L. and Orth, K. (2009). AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323(5911): 269-272.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Truttmann, M. C. and Ploegh, H. L. (2017). In vitro AMPylation Assays Using Purified, Recombinant Proteins. Bio-protocol 7(14): e2416. DOI: 10.21769/BioProtoc.2416.
Category
Biochemistry > Protein > Modification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










