- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
RNase Sensitivity Screening for Nuclear Bodies with RNA Scaffolds in Mammalian Cells
Published: Vol 7, Iss 8, Apr 20, 2017 DOI: 10.21769/BioProtoc.2232 Views: 10808
Reviewed by: Gal HaimovichAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Studying Cellular Focal Adhesion Parameters with Imaging and MATLAB Analysis
Ling-Yea Yu [...] Feng-Chiao Tsai
Nov 5, 2023 1879 Views

Automated Layer Analysis (ALAn): An Image Analysis Tool for the Unbiased Characterization of Mammalian Epithelial Architecture in Culture
Christian Cammarota [...] Tara M. Finegan
Apr 20, 2024 4222 Views

Accurate Identification of Cell Cycle Stages in RPE1 Cells Using the ImmunoCellCycle-ID Method
Syon Reddy [...] Aussie Suzuki
Aug 5, 2025 1870 Views
Abstract
The mammalian cell nucleus is highly organized and contains membraneless nuclear bodies (NBs) characterized by distinct resident factors. The NBs are thought to serve as sites for biogenesis and storage of certain RNA and protein factors as well as assembly of ribonucleoprotein complexes. Some NBs are formed with architectural RNAs (arcRNAs) as their structural scaffolds and additional NBs likely remain unidentified in mammalian cells. Here, we describe an experimental protocol to search for new NBs built on certain arcRNAs. RNase-sensitive NBs were identified by monitoring nuclear foci visualized by tagging thousands of human cDNA products.
Keywords: Nuclear bodiesBackground
The mammalian cell nucleus is highly organized and composed of multiple distinct substructures called nuclear bodies (NBs). So far ~15 NBs have been identified as subnuclear membraneless granular structures containing various proteins and RNA factors, many of which function as sites of biogenesis, maturation, storage, and sequestration of specific RNAs, proteins, and/or ribonucleoprotein (RNP) complexes (Mao et al., 2011; Sleeman and Trinkle-Mulcahy, 2014) (Table 1).
Table 1. Nuclear bodies in mammalian cells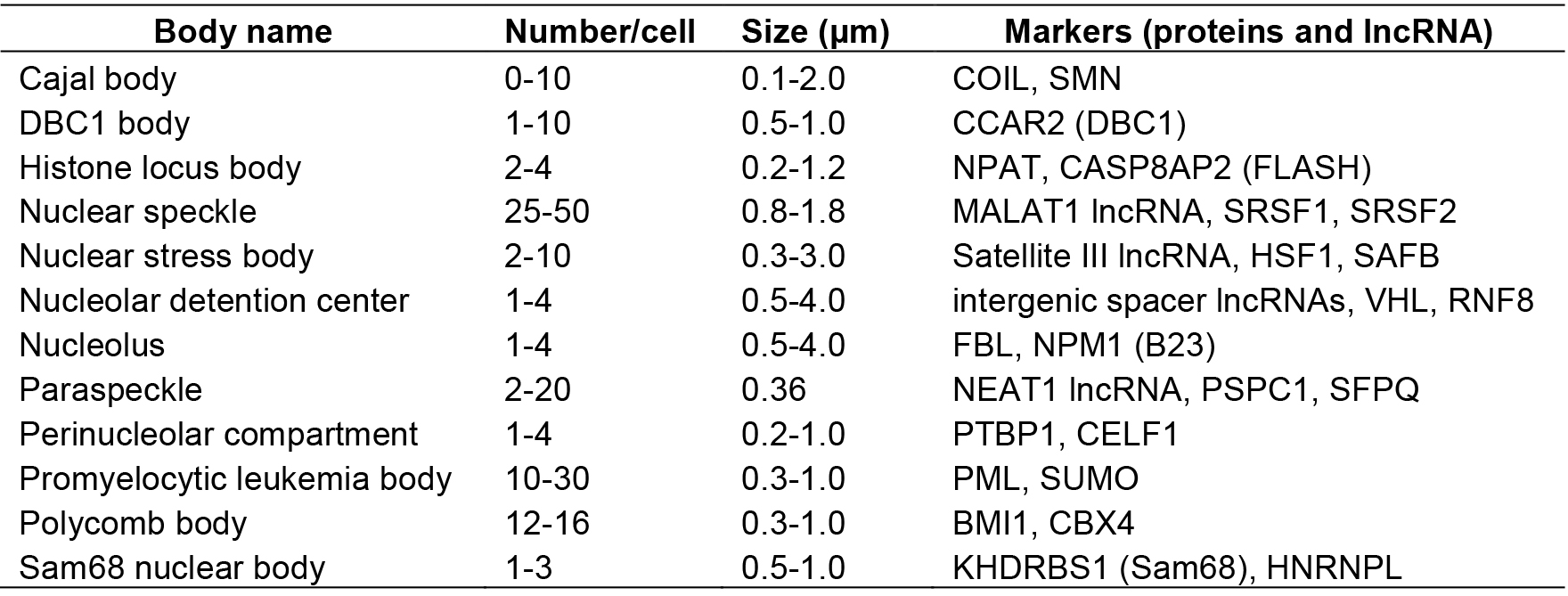
Some NBs are constructed on specific long noncoding RNAs (lncRNAs) called architectural RNAs (arcRNAs), which are defined as structural core of NBs (Chujo et al., 2016). The arcRNA-dependent NBs are composed of numerous RNA-binding proteins that interact with the arcRNAs. The most remarkable example is the paraspeckle, which is composed of several characteristic RNA-binding proteins (Fox et al., 2002; Prasanth et al., 2005). RNase treatment disintegrates the paraspeckle structure (Fox et al., 2005). Nuclear paraspeckle assembly transcript 1 (NEAT1), a lncRNA, localizes exclusively to paraspeckles and acts as an arcRNA of these massive RNP complexes (Chen and Carmichael, 2009; Clemson et al., 2009; Sasaki et al., 2009; Sunwoo et al., 2009).
Presently, two lncRNAs are classified as arcRNAs in addition to NEAT1, namely, intergenic spacer lncRNAs for nucleolar detention center (Audas et al., 2012) and human satellite III lncRNA for nuclear stress body (Biamonti and Vourc’h, 2010) (Table 1). It is expected that additional arcRNAs remain to be characterized in mammalian cells. Here, we describe a novel method called ‘RNase sensitivity screening’ to identify novel arcRNA-dependent NBs by screening for nuclear foci whose structures are disintegrated by RNase treatment. This method employed a Venus-tagged human full-length (FLJ) cDNA library (32,651 clones), which was originally constructed during the NEDO full-length human cDNA sequencing project in Japan (FLJ-PJ), and obtained 571 cDNA clones whose products (463 proteins) localize in certain nuclear foci (Hirose and Goshima, 2015; Naganuma et al., 2012). We explored whether the respective nuclear focus was abolished or diffused upon RNase treatment after cell permeabilization to select candidate RNase-sensitive nuclear foci that potentially contain arcRNAs (Figure 1). We identified 32 Venus-tagged proteins that required RNA for their localization in distinct nuclear foci (Mannen et al., 2016). Immunostaining of the corresponding endogenous proteins confirmed that the Sam68 nuclear body (SNB) was an RNase-sensitive NB. In the following protocol, we describe the detailed procedure of the RNase sensitivity screening. This protocol is for screening for RNase-sensitive NBs under normal conditions in HeLa cells, but it should be applicable to other cell lines under various conditions.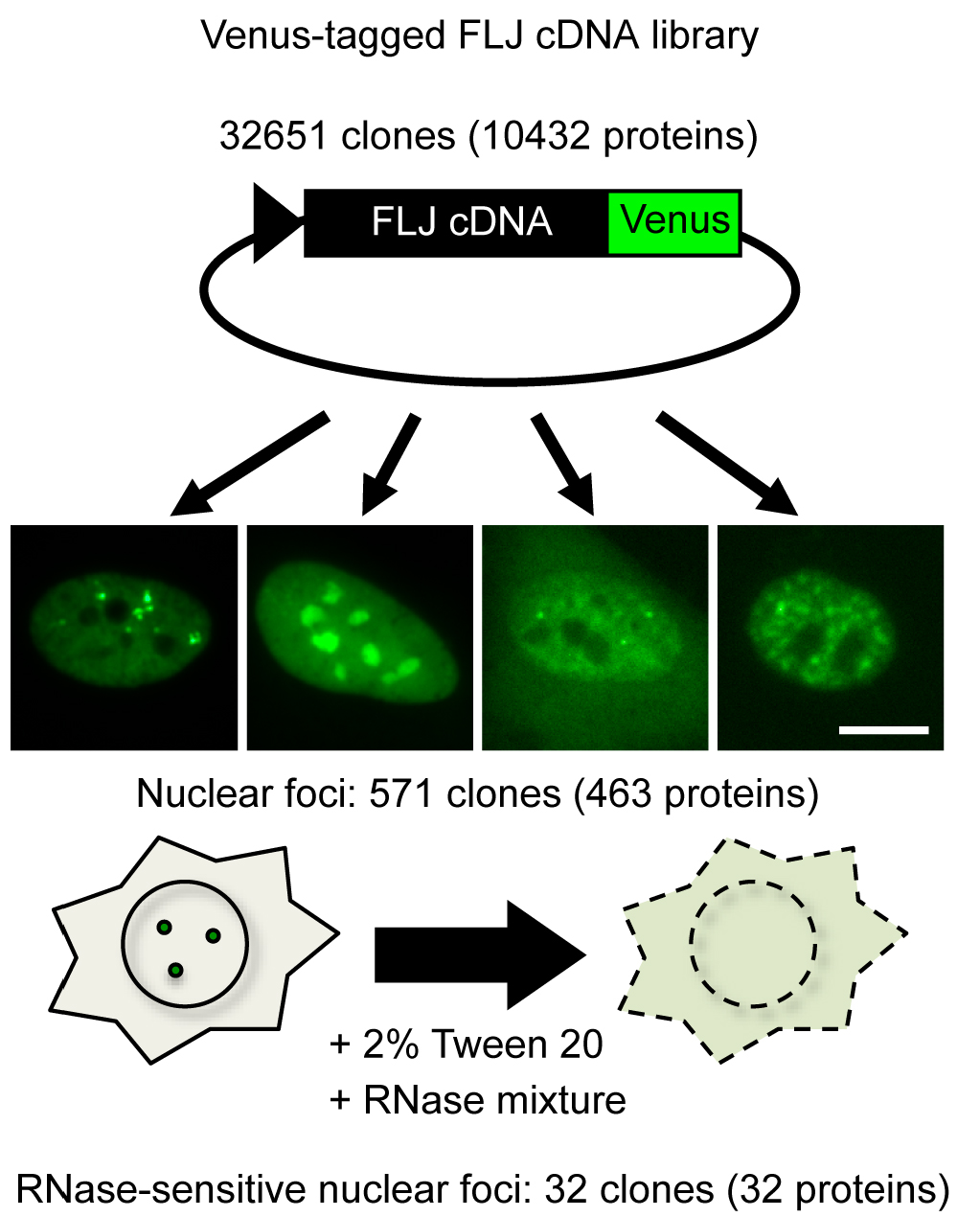
Figure 1. Outline of RNase sensitivity screening of NBs. Venus-tagged human FLJ cDNA clones were transfected into HeLa cells. cDNA clones whose products localized to certain nuclear foci were selected (571 clones). Subsequently, the RNase sensitivity of the nuclear foci labeled by Venus was investigated (32 clones). To this end, the cells were permeabilized with 2% Tween 20, followed by treatment with an RNase mixture. Bar = 10 μm.
Materials and Reagents
Note: Prepare all solutions using ultrapure water (prepared by purifying deionized water to attain a sensitivity of 18.2 MΩ cm at 25 °C) and analytical grade reagents.
- Preparation of collagen IV-coated plate
- Pipette tips (Labcon, catalog number: 1030-260-000 )
- 96-well glass bottom microplate (IWAKI, catalog number: 12-017-006 )
- Sodium hydroxide (NaOH) (Wako Pure Chemical Industries, catalog number: 190-14565 )
- Phosphate Buffered Salts (PBS) (Takara Bio, catalog number: T900 )
- Hydrochloric acid (HCl) (Wako Pure Chemical Industries, catalog number: 080-01066 )
- Type IV collagen solution (Life Laboratory Company, catalog number: LL-10043 )
- Collagen solution (see Note 1; see Recipes)
- Pipette tips (Labcon, catalog number: 1030-260-000 )
- Cell culture
- HeLa (Human cervical cancer) cells (see Note 2)
- 10 cm dish
- MEM medium (Thermo Fisher Scientific, GibcoTM, catalog number: 11095080 ) (see Note 3)
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 26140079 )
- Trypsin-EDTA (Nacalai Tesque, catalog number: 32778-34 )
- HeLa (Human cervical cancer) cells (see Note 2)
- Administration of plasmids with transfection reagent
- 96-well U-bottom plate (Corning, Falcon®, catalog number: 351177 )
- Venus-tagged human FLJ cDNA clone plasmids expressing proteins localized to certain nuclear foci (see Note 4)
- TransIT-LT1 reagent (Mirus Bio, catalog number: MIR2300 )
- Opti-MEM I reduced serum medium (Thermo Fisher Scientific, GibcoTM, catalog number: 31985070 )
- 96-well U-bottom plate (Corning, Falcon®, catalog number: 351177 )
- RNase treatment
- 96-well plate sealing film (Thermo Fisher Scientific, InvitrogenTM, catalog number: 12261012 )
- 0.45 μm filter
- RiboShredder RNase Blend (Epicentre, catalog number: RS12500 )
- Polyoxyethylene sorbitan monolaurate (Tween 20) (Nacalai Tesque, catalog number: 28353-85 )
- 2-Amino-2-hydroxymethyl-1,3-propanediol (Tris) (Wako Pure Chemical Industries, catalog number: 011-20095 )
- Magnesium chloride (MgCl2) (Nacalai Tesque, catalog number: 20909-55 )
- Ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid (EGTA) (Nacalai Tesque, catalog number: 08907-42 )
- cOmplete, EDTA-free protease inhibitor cocktail (Roche Diagnostics, catalog number: 11873580001 )
- Paraformaldehyde (PFA) (EMD Millipore, catalog number: 1.04005.1000 )
- DAPI (Sigma-Aldrich, catalog number: D9542 )
- Pyronin Y (Sigma-Aldrich, catalog number: P9172 )
- 1 M Tris-HCl, pH 7.4 (see Recipes)
- Permeabilization buffer (see Recipes)
- 4% PFA (see Note 5; see Recipes)
- DAPI solution (see Recipes)
- Pyronin Y solution (see Recipes)
- 96-well plate sealing film (Thermo Fisher Scientific, InvitrogenTM, catalog number: 12261012 )
Equipment
- Multichannel pipette L8 x 200 (Gilson, catalog number: FA10011 )
- Water bath (Fine, catalog number: FWB-21B )
- Centrifuge (KUBOTA, catalog number: 040-000 )
- Hemocytometer (SANSYO, SLGC, catalog number: A103 )
- Reservoir (BM Equipment, catalog number: BM-0850-2 )
- Carbon dioxide (CO2) incubator (SANYO, catalog number: MCO-18AIC UV )
- IN Cell Analyzer 1000 (GE Healthcare, model: IN Cell Analyzer 1000 ) equipped with a Plan Fluor ELWD 40x/0.6 objective lens (Nikon Instruments, model: CFI Plan Fluor 40x ). Excitation filters for DAPI (D360/40x) and Venus (S475/20x) and emission filters for DAPI (HQ460/40M) and Venus (HD535/50M) were used. Acquisition and processing of the images were done using IN Cell Analyzer 1000 Software (GE Healthcare, version 3.0)
Software
- IN Cell Analyzer 1000 Software (GE Healthcare, version 3.0)
Procedure
- Preparation of collagen-coated plate
- Wash each well of 96-well glass bottom microplates with 100 μl 0.2 N NaOH at room temperature for 30 min.
- Rinse each well with 100 μl PBS and then 100 μl distilled water.
- Coat the glass bottom of each well with 100 μl collagen solution at room temperature for 1 h (see Note 6).
- Wash each well with 100 μl PBS twice.
- Remove PBS completely from each well by pipetting. The plates can be stored at 4 °C for 1 week.
- Wash each well of 96-well glass bottom microplates with 100 μl 0.2 N NaOH at room temperature for 30 min.
- Cell culture
- Thaw frozen cell stocks in a water bath at 37 °C, and retrieve the cells by low speed centrifugation (220 x g, 5 min). Remove the supernatant by pipetting.
- Gently resuspend the cell pellet in 10 ml growth medium (e.g., MEM medium supplemented with 10% FBS) without antibiotics.
- Culture the cells in growth medium in a humidified incubator with 5% CO2 until 80% confluency is reached.
- For plasmid transfection, seed the cells into a 10 cm dish 1 day prior to use (see Note 7).
- Thaw frozen cell stocks in a water bath at 37 °C, and retrieve the cells by low speed centrifugation (220 x g, 5 min). Remove the supernatant by pipetting.
- Administration of plasmids with transfection reagent
- Seed 6 x 103 cells into 100 μl growth medium in each well of a collagen-coated 96-well plate 1 day prior to transfection. The cells should be used for transfection when they have reached 30-50% confluency.
- Add 100 ng Venus-tagged FLJ cDNA clone plasmids to 50 μl Opti-MEM I reduced medium in each well of a 96-well U-bottom plate.
- Add 0.3 μl TransIT-LT1 reagent to 50 μl Opti-MEM I reduced medium in each well of a separate 96-well U-bottom plate.
- Mix the solutions prepared in steps C2 and C3 and incubate at room temperature for 15-30 min to form the plasmid-TransIT-LT1 complexes.
- Add the resultant plasmid-TransIT-LT1 complexes (ca. 100 μl) directly to the growth medium in each well of the 96-well plate prepared in step C1.
- Culture the cells for 24 h in the incubator with 5% CO2 (see Note 8).
- Seed 6 x 103 cells into 100 μl growth medium in each well of a collagen-coated 96-well plate 1 day prior to transfection. The cells should be used for transfection when they have reached 30-50% confluency.
- RNase treatment
- Remove the culture medium by pipetting and wash the cells once with 100 μl PBS.
- Wash the cells once with 100 μl permeabilization buffer and then permeabilize the cells with 100 μl 2% Tween 20-permeabilization buffer at room temperature for 10 min (see Note 9).
- Wash the cells once with 100 μl permeabilization buffer.
- Wash the cells once with 100 μl PBS and degrade RNAs with 100 μl 50 U/ml RiboShredder RNase Blend-PBS at room temperature for 20 min (see Note 10).
- Wash the cells twice with 100 μl PBS.
- Fix the cells with 100 μl 4% PFA at room temperature for 15 min.
- Wash the cells three times with 100 μl PBS for 5 min each.
- Stain the DNA with 100 μl DAPI solution for 1 min.
- Wash the cells three times with 100 μl PBS.
- (Optional, see Note 11) Stain the RNA with 100 μl Pyronin Y solution for 15 sec.
- (Optional) Wash the cells three times with 100 μl PBS.
- Add 100 μl PBS to each well and seal the wells with a 96-well plate sealing film.
- Visualize Venus signals and detect nuclear foci in the samples using the IN Cell Analyzer 1000 (Figure 2). The images were acquired under 400x magnification, with automatically optimized exposure times and hardware autofocus. Images of 25 different fields were acquired from each well. (see Note 12)
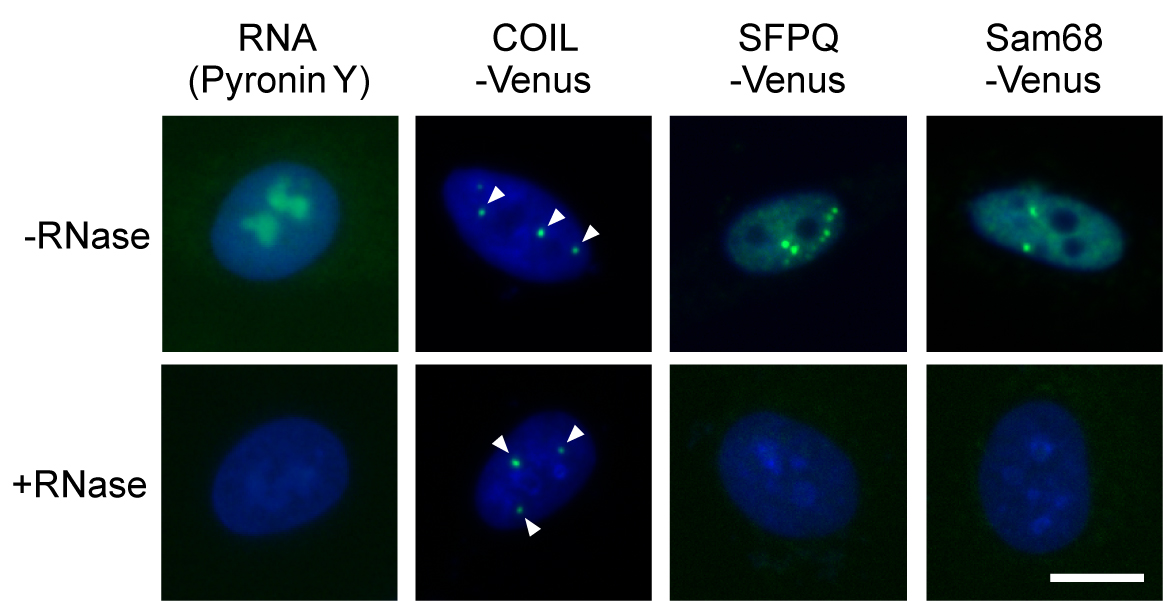
Figure 2. Images of RNase sensitivity screening of NBs. Effect of RNase treatment was confirmed by visualizing cellular RNAs with Pyronin Y staining, and by monitoring the persistence of the RNase-resistant Cajal bodies (marked by COIL-Venus) and disappearance of the RNase-sensitive paraspeckles (marked by SFPQ-Venus). Sam68 nuclear body is a newly identified RNase-sensitive NB. Arrowheads indicate nuclear foci. DNA was stained with DAPI (blue). Bar = 10 μm.
- Remove the culture medium by pipetting and wash the cells once with 100 μl PBS.
Data analysis
Cell numbers (n > 100) were determined by counting the outlined DAPI positive oval area as the nucleus using IN Cell Analyzer 1000 Software. The number of nuclear foci-positive cells was manually counted. RNase sensitivity is measured by the ratio of the number of nuclear foci-positive cells in RNase-treated cells relative to the number of those in the cells without RNase treatment.
Notes
- This solution should be prepared at the time of use.
- Although the protocol described here is for HeLa cells, it may be applicable to other cultured cell lines (e.g., HCT116, U2OS, and NIH3T3). These cell lines are available from several cell stock centers, such as the American Type Culture Collection.
- Culture medium should be properly chosen for the cell lines used.
- The list of FLJ clones expressing the Venus-tagged proteins that localized to certain nuclear foci is available at the following website: http://hgpd.lifesciencedb.jp/sys_info/download.html. The localization images of the Venus-tagged human cDNA products are accessible at the Human Gene and Protein Database (HGPD). The original Gateway Entry clones are available from the Biological Resource Center, National Institute of Technology and Evaluation (NBRC). We used two Venus-tagged proteins as controls: SFPQ-Venus (a marker of the paraspeckle, an RNase-sensitive NB) and COIL-Venus (a marker of the Cajal body, an RNase-resistant NB).
- This solution should be prepared at the time of use.
- Other coating reagents such as cationic polymers (e.g., poly-L-lysine) or matrix proteins (e.g., laminin, fibronectin) can be selected according to the cells used.
- The number of cell passages should be less than five.
- Transfection conditions should be optimized by varying the concentration of TransIT-LT1 reagent.
- Permeabilization conditions should be optimized by monitoring the control Venus-tagged proteins (Note 4) and Pyronin Y staining. The concentration of Tween 20 should be optimized for each cell line used. It should be noted that treatment with higher concentrations of Tween 20 often results in disappearance of Venus signals in the cells.
- RNase conditions should be optimized by monitoring the control Venus-tagged proteins (Note 4) and Pyronin Y staining. We always compare two samples with and without RNase treatment for one cDNA clone.
- Effects of RNase treatment are confirmed by staining of cellular RNAs with Pyronin Y.
- For final judgment of localization sites of the selected cDNA products, both N-terminally and C-terminally Venus-tagged proteins should be confirmed to consistently localize in identical NBs. Furthermore, the localization of the corresponding endogenous proteins should be confirmed by labeling with specific antibodies.
Recipes
- Collagen solution
Dilute 8.6 μl/ml collagen in 0.05 N HCl - 1 M Tris-HCl, pH 7.4 (1 L)
Dilute 121.1 g Tris in ultrapure water and adjust pH to 7.4 with HCl - Permeabilization buffer
20 mM Tris-HCl, pH 7.4
5 mM MgCl2
0.5 mM EGTA
1x cOmplete, EDTA-free protease inhibitor cocktail - 4% PFA
Add 0.4 g PFA to PBS and adjust the volume to 10 ml
Shake vigorously at 55 °C until completely dissolved and filtrate through a 0.45 μm filter - DAPI solution
Add 10 mg DAPI to ultrapure water and adjust the volume to 1 ml (10 mg/ml DAPI stock)
Store at -20 °C
Before using, dilute 1 μl 10 mg/ml DAPI stock in 10 ml PBS (1 μg/ml DAPI) - Pyronin Y solution
Add 3 mg Pyronin Y to ultrapure water and adjust the volume to 10 ml (1 mM Pyronin Y stock)
Store at -20 °C
Before using, dilute 1,000 times with PBS (1 μM Pyronin Y)
Acknowledgments
This protocol was adapted from our recently published work (Mannen et al., 2016). We thank N. Goshima, Y. Kawamura, and H. Mochizuki at the National Institute of Advanced Industrial Science and Technology for their support in the preparation of the Venus-tagged cDNA library. This research was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (26113002 and 26291001).
References
- Audas, T. E., Jacob, M. D. and Lee, S. (2012). Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 45(2): 147-157.
- Biamonti, G. and Vourc'h, C. (2010). Nuclear stress bodies. Cold Spring Harb Perspect Biol 2(6): a000695.
- Chen, L. L. and Carmichael, G. G. (2009). Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35(4): 467-478.
- Chujo, T., Yamazaki, T. and Hirose, T. (2016). Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Biophys Acta 1859(1): 139-146.
- Clemson, C. M., Hutchinson, J. N., Sara, S. A., Ensminger, A. W., Fox, A. H., Chess, A. and Lawrence, J. B. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33(6): 717-726.
- Fox, A. H., Bond, C. S. and Lamond, A. I. (2005). P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 16(11): 5304-5315.
- Fox, A. H., Lam, Y. W., Leung, A. K., Lyon, C. E., Andersen, J., Mann, M. and Lamond, A. I. (2002). Paraspeckles: a novel nuclear domain. Curr Biol 12(1): 13-25.
- Hirose, T. and Goshima, N. (2015). Genome-wide co-localization screening of nuclear body components using a fluorescently tagged FLJ cDNA clone library. Methods Mol Biol 1262: 155-163.
- Mannen, T., Yamashita, S., Tomita, K., Goshima, N. and Hirose, T. (2016). The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. J Cell Biol 214(1): 45-59.
- Mao, Y. S., Zhang, B. and Spector, D. L. (2011). Biogenesis and function of nuclear bodies. Trends Genet 27(8): 295-306.
- Naganuma, T., Nakagawa, S., Tanigawa, A., Sasaki, Y. F., Goshima, N. and Hirose, T. (2012). Alternative 3’end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J 31(20): 4020-4034.
- Prasanth, K. V., Prasanth, S. G., Xuan, Z., Hearn, S., Freier, S. M., Bennett, C. F., Zhang, M. Q. and Spector, D. L. (2005). Regulating gene expression through RNA nuclear retention. Cell 123(2): 249-263.
- Sasaki, Y. T., Ideue, T., Sano, M., Mituyama, T. and Hirose, T. (2009). MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A 106(8): 2525-2530.
- Sleeman, J. E. and Trinkle-Mulcahy, L. (2014). Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr Opin Cell Biol 28: 76-83.
- Sunwoo, H., Dinger, M. E., Wilusz, J. E., Amaral, P. P., Mattick, J. S. and Spector, D. L. (2009). MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19(3): 347-359.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Mannen, T. and Hirose, T. (2017). RNase Sensitivity Screening for Nuclear Bodies with RNA Scaffolds in Mammalian Cells. Bio-protocol 7(8): e2232. DOI: 10.21769/BioProtoc.2232.
-
Mannen, T., Yamashita, S., Tomita, K., Goshima, N. and Hirose, T. (2016). The Sam68 nuclear body is composed of two RNase-sensitive substructures joined by the adaptor HNRNPL. J Cell Biol 214(1): 45-59.
Category
Cell Biology > Cell imaging > Fixed-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








