- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vivo Mitophagy Monitoring in Caenorhabditis elegans to Determine Mitochondrial Homeostasis
Published: Vol 7, Iss 7, Apr 5, 2017 DOI: 10.21769/BioProtoc.2215 Views: 11444
Reviewed by: Jyotiska ChaudhuriPia GiovannelliAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Simultaneous Monitoring Cytoplasmic Calcium Ion and Cell Surface Phosphatidylserine in the Necrotic Touch Neurons of Caenorhabditis elegans
Yoshitaka Furuta and Zheng Zhou
Oct 20, 2021 2623 Views

Live-cell Imaging and Analysis of Germline Stem Cell Mitosis in Caenorhabditis elegans
Réda M. Zellag [...] Abigail R. Gerhold
Jan 5, 2022 4694 Views

SunTag-Based Single-Molecule Translation Imaging in Caenorhabditis elegans
Elise van der Salm [...] Suzan Ruijtenberg
Oct 20, 2025 2281 Views
Abstract
Perturbation of mitochondrial function is a major hallmark of several pathological conditions and ageing, underlining the essential role of fine-tuned mitochondrial activity (Lopez-Otin et al., 2013). Mitochondrial selective autophagy, known as mitophagy, mediates the removal of dysfunctional and/or superfluous organelles, preserving cellular and organismal homeostasis (Palikaras and Tavernarakis, 2014; Pickrell and Youle, 2015; Scheibye-Knudsen et al., 2015). In this protocol, we describe a method for assessing mitophagy in the nematode Caenorhabditis elegans.
Keywords: AgeingBackground
Mitochondria are characterized as cellular powerhouses of eukaryotic cells, since they are the major energy providers through oxidative phosphorylation (OXPHOS) and ATP generation. Moreover, their pivotal role in cellular homeostasis is highlighted by their contribution in the regulation of several fundamental cellular processes including calcium buffering, metabolite synthesis and apoptosis, among others. Deregulation of mitochondrial function is associated with the onset of several pathological conditions including ageing and age-related neurodegenerative diseases (Vafai and Mootha, 2012; Palikaras and Tavernarakis, 2014). Thus, eukaryotic organisms have evolved several complex and highly specialized molecular pathways to guard energy homeostasis (Pickrell and Youle, 2015; Scheibye-Knudsen et al., 2015). Mitophagy is a selective type of autophagy promoting the elimination of impaired mitochondria, and the major degradation pathway by which cells regulate mitochondrial content in response to intracellular and environmental signals (Palikaras et al., 2015; Schiavi et al., 2015; Fang et al., 2016). In this protocol, we describe two methods for monitoring mitophagy in C. elegans. We developed two composites, in vivo imaging systems to asses mitophagy based, first, on the Rosella biosensor (Rosado et al., 2008), which combines a fast-maturing pH-insensitive DsRed fused to a pH-sensitive GFP variant, and second, on a custom, dual-fluorescence reporter system that involves a mitochondria-targeted GFP, together with the autophagosomal marker LGG-1/LC3 fused to DsRed. These protocols facilitate non-invasive monitoring of mitophagy in live specimens.
Materials and Reagents
- Greiner Petri dishes (60 x 15 mm) (Greiner Bio One International, catalog number: 628161 )
- Microscope slides 75 x 25 x 1 mm (Marienfeld-Superior, catalog number: 10 006 12 )
- Microscope cover glass 18 x 18 mm (Marienfeld-Superior, catalog number: 01 010 30 )
- Use the following transgenic nematodes to monitor mitophagy: IR1631: N2;Ex003 [pmyo-3TOMM-20::Rosella; pRF4], R1284: N2;Is [pmyo-3mtGFP];Ex011 [plgg-1DsRed::LGG-1; pmyo-2GFP])
- Escherichia coli OP50 strain (obtained from the Caenorhabditis Genetics Center)
- 70% of EtOH
- Potassium dihydrogen phosphate (KH2PO4) (EMD Millipore, catalog number: 104873 )
- di-Potassium hydrogen phosphate (K2HPO4) (EMD Millipore, catalog number: 137010 )
- Sodium chloride (NaCl) (EMD Millipore, catalog number: 106404 )
- Peptone (BD, BactoTM, catalog number: 211677 )
- Streptomycin (Sigma-Aldrich, catalog number: S6501 )
- Agar (Sigma-Aldrich, catalog number: 05040 )
- Cholesterol stock solution (SERVA Electrophoresis, catalog number: 17101.01 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C5080 )
- Magnesium sulfate (MgSO4) (Sigma-Aldrich, catalog number: M7506 )
- Nystatin stock solution (Sigma-Aldrich, catalog number: N3503 )
- di-Sodium hydrogen phosphate (Na2HPO4) (EMD Millipore, catalog number: 106586 )
- Levamisole (Sigma-Aldrich, catalog number: L9756 )
- Paraquat (Sigma-Aldrich, catalog number: 856177 )
- Carbonyl cyanide m-chlorophenylhydrazone (CCCP) (Sigma-Aldrich, catalog number: C2759 )
- Dimethyl sulfoxide cell culture grade BC (DMSO) (AppliChem, catalog number: A3672,0250 )
- Phosphate buffer (1 M; sterile, see Recipes)
- Nematode growth medium (NGM) agar plates (see Recipes)
- M9 buffer (see Recipes)
- Levamisole (0.5 M, see Recipes)
- M9-levamisole solution (20 mM solution, see Recipes)
- Paraquat (0.5 M, see Recipes)
- Carbonyl cyanide m-chlorophenylhydrazone (49 mM; CCCP, see Recipes)
Equipment
- UV crosslinker (Vilber Lourmat, model: BIO-LINK – BLX-E365 )
- Zeiss AxioImager Z2 epifluorescence microscope (Zeiss, model: Zeiss AxioImager Z2 )
- Olympus DP71 CCD camera (Olympus, model: Olympus DP71 )
- Zeiss AxioObserver Z1 confocal microscope (Zeiss, model: Zeiss AxioObserver Z1 )
- Dissecting stereomicroscope (Olympus, model: SMZ645 )
- Incubators for stable temperature (AQUA®LYTIC incubator 20 °C)
Software
- Olympus CELL-A software
- Zeiss ZEN 2012 software
- Image J (https://imagej.nih.gov/ij/)
- Microsoft Office 2011 Excel (Microsoft Corporation, Redmond, USA)
- GraphPad Prism software package (GraphPad Software Inc., San Diego, USA)
Procedure
- Growth and synchronization of nematode population
- Select L4 larvae of transgenic animals, which express mitochondria-targeted Rosella (mtRosella) or co-express mitochondria-targeted GFP (mtGFP) together with the autophagosomal marker LGG-1 fused with DsRed in body wall muscle cells, on a freshly E. coli (OP50) seeded NGM plate. Use at least three plates for each nematode strain.
- Incubate the nematodes at the standard temperature of 20 °C.
- Four days later the plates contain mixed animals population.
- Synchronize nematodes by picking L4 transgenic larvae and transfer them onto separate freshly E. coli (OP50) seeded plates (Palikaras, K. and Tavernarakis, N. [2016]. Intracellular Assessment of ATP Levels in Caenorhabditis elegans. Bio-protocol 6(23): e2048; Palikaras, K. and Tavernarakis, N. [2016]. Measuring Oxygen Consumption Rate in Caenorhabditis elegans. Bio-protocol 6(23): e2049).
- Add 20 L4 larvae per plate. For each experimental condition, use at least five plates (see Note 1).
- Select L4 larvae of transgenic animals, which express mitochondria-targeted Rosella (mtRosella) or co-express mitochondria-targeted GFP (mtGFP) together with the autophagosomal marker LGG-1 fused with DsRed in body wall muscle cells, on a freshly E. coli (OP50) seeded NGM plate. Use at least three plates for each nematode strain.
- Oxidative and mitochondrial stress assay
- Kill E. coli (OP50) bacteria seeded on NGM plates by using a UV crosslinker (UV irradiation for 15 min; 0.5 J).
- Add paraquat or CCCP to the top of seeded NGM plates at 8 mM and 15 μM final concentrations in the total agar volume respectively. Paraquat and CCCP are well-known inducers of mitophagy in mammalian cells (Narendra et al., 2010).
- Gently swirl the plates and allow each drug to spread to the entire surface.
- Let the plates dry at room temperature.
- Transfer 2- or 4-day-old of adulthood adult transgenic animals on paraquat- or CCCP-containing plates.
- Incubate the transgenic animals at 20 °C.
- Upon two days of exposure to each drug prepare the nematodes for microscopic examination.
- Kill E. coli (OP50) bacteria seeded on NGM plates by using a UV crosslinker (UV irradiation for 15 min; 0.5 J).
- Mounting nematodes for imaging
- Add a droplet of 10 μl M9-levamisole buffer (20 mM final concentration, see Note 2) on 2% agarose pad (see Note 3).
- Collect transgenic animals with an eyebrow/eyelash hair and place them in M9-levamisole droplet immobilizing transgenic animals for imaging (see Note 4).
- Gently place a coverslip on the top press. Samples are ready for microscopic examination with either a Zeiss AxioImager Z2 epifluorescence microscope or a Zeiss AxioObserver Z1 confocal microscope.
- Add a droplet of 10 μl M9-levamisole buffer (20 mM final concentration, see Note 2) on 2% agarose pad (see Note 3).
- Imaging of transgenic nematodes
- Capture single transgenic animals or single body wall muscle cells using a camera attached to the microscope.
- Acquire (a) fluorescent images of whole transgenic nematodes expressing mtRosella in body wall muscle cells by using Zeiss AxioImager Z2 epifluorescence microscope or perform (b) z-stack method of an entire single body wall muscle cell expressing mtGFP together with autophagosomal marker DsRed::LGG-1 by using Zeiss AxioObserver Z1 confocal microscope.
- Imaging parameters such as microscope and camera settings (lens and magnifier used, filters exposure time, resolution, laser intensity, gain etc.) should be documented and kept the same during the imaging process.
- Save collected images from each method (a or b) and proceed to data analysis.
- Capture single transgenic animals or single body wall muscle cells using a camera attached to the microscope.
- Analyze data from imaging process
- Process images acquired from (a) method with ImageJ software to measure the average pixel intensity values and total area for each fluorescent image of transgenic worm. Focus on body wall muscle cells of the head region avoiding intestinal autofluorescence (Figures 1 and 2; see Note 5). To analyze the area of interest manually:
- Select the ‘split channel’ command via the ‘image’ and ‘colour’ drop-down menu to convert images.
- Use the ‘freehand selection’ tool to enclose the fluorescent area.
- Select the ‘measurement’ command via the ‘analyze’ drop-down menu to perform pixel intensity analysis.
- Normalize pixel intensity values by dividing with the selected area values by using the Microsoft Office 2011 Excel software package (Microsoft Corporation, Redmond, USA).
- Upon normalization calculate GFP to DsRed ratio.
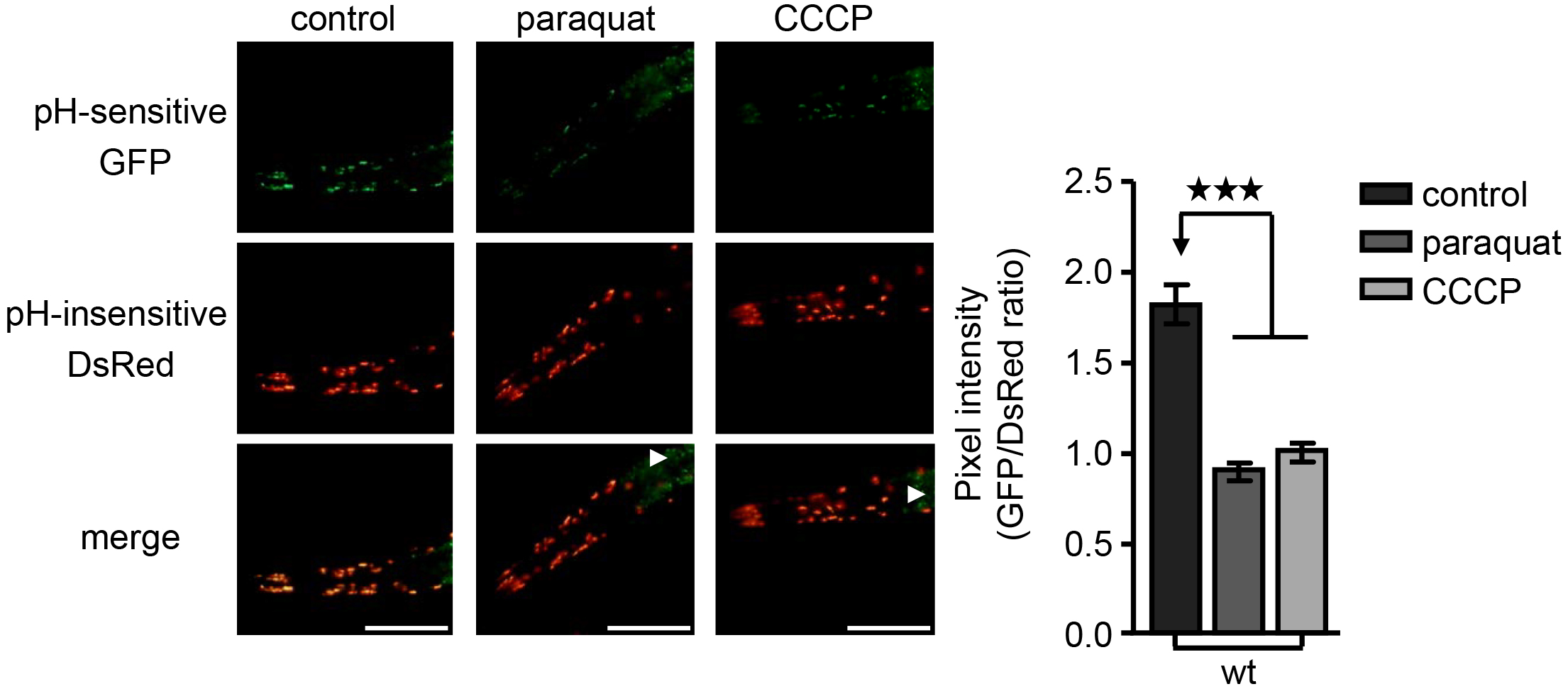
Figure 1. Mitophagy induction upon oxidative and mitochondrial stress. Transgenic nematodes expressing mtRosella in body wall muscle cells, were exposed to paraquat or CCCP. Mitophagy induction is signified by the reduction of the ratio between pH-sensitive GFP to pH-insensitive DsRed (n = 100; ***P < 0.001; one-way ANOVA). Arrowheads point out intestinal autofluorescence. Size bars denote 20 μm. Images were acquired using a 10x objective lens. Error bars denote SEM values.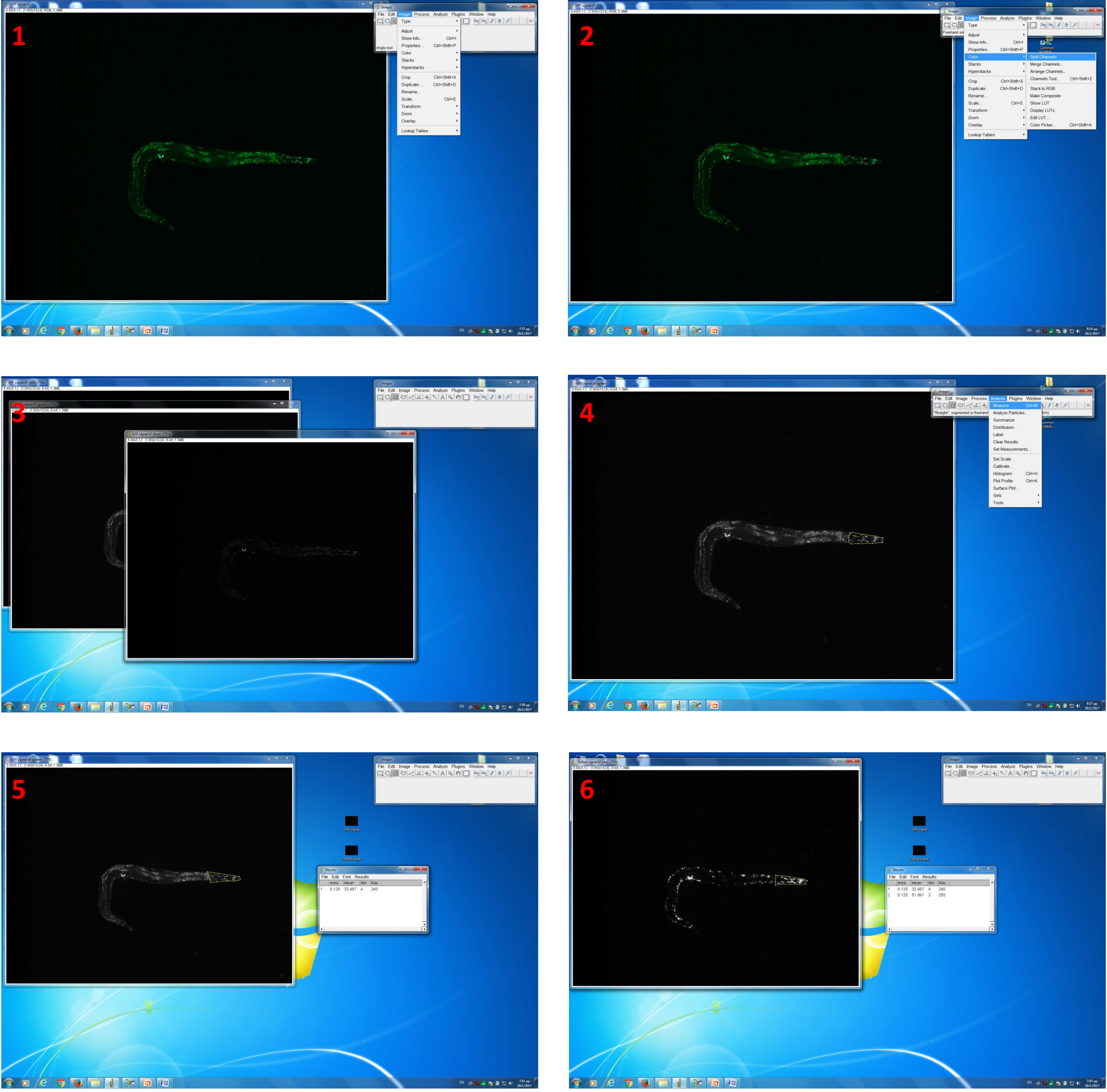
Figure 2. Image analysis by using ImageJ software. 1. Open an acquired GFP signal-image with ImageJ software; 2. Select split channel’ command via the ‘image’ and ‘colour’ drop-down menu to convert images; 3. Keep ‘green channel’ image; 4. Use the ‘freehabnd selection’ tool to enclose the fluorescent area (head region); 5. Select the ‘measurement’ command via the ‘analyze’ drop-down menu to perform pixel intensity analysis; 6. Perform the same analysis pathway with an acquired DsRed-signal image.
- Select the ‘split channel’ command via the ‘image’ and ‘colour’ drop-down menu to convert images.
- Process images collected from (b) method with Zeiss ZEN 2012 software to find mitochondria engulfed by autophagosomes, known as mitoautophagosomes.
- Determine mitoautophagosomes number by manually counting the co-localization events between mitochondrial (mtGFP) and autophagosomal marker (DsRed::LGG-1) displayed in each stack of body wall muscle cell (Figure 3).
- Analyze the data by using the Microsoft Office 2011 Excel software package (Microsoft Corporation, Redmond, USA).
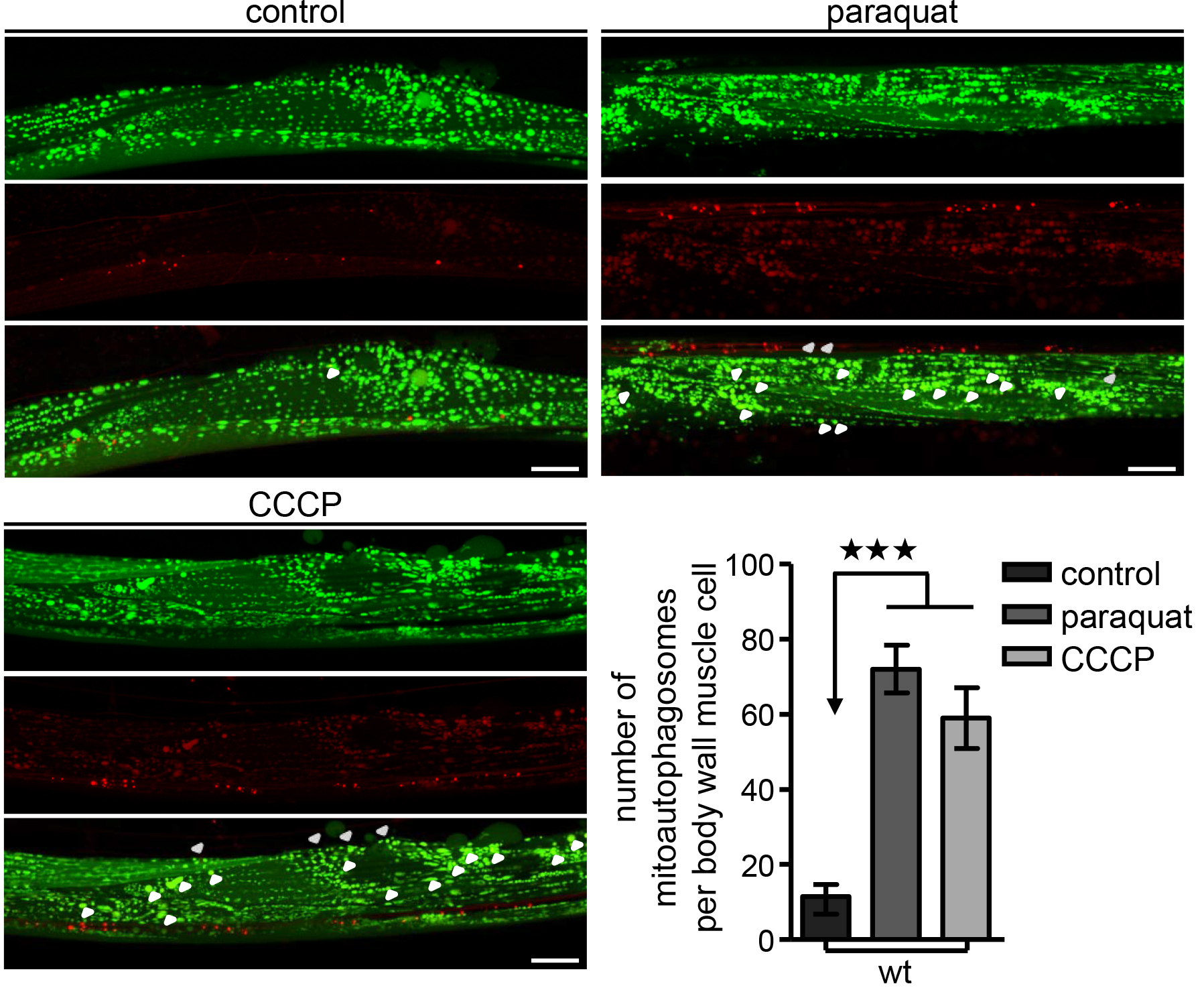
Figure 3. Monitor mitoautophagosomes formation in vivo. Transgenic nematodes co-expressing a mitochondria-targeted GFP (mtGFP) in body wall muscle cells together with the autophagosomal protein LGG-1 fused with DsRed, were treated with paraquat or CCCP. Mitophagy stimulation is signified by co-localization of GFP and DsRed signals (for each group of images mitochondria are shown in green on top, autophagosomes in red below, with a merged image at the bottom). Increased number of mitoautophagosomes upon oxidative and mitochondrial stress (n = 50; ***P < 0.001; one-way ANOVA). Size bars denote 20 μm. Images were acquired using a 40x objective lens. Error bars denote SEM values.
- Determine mitoautophagosomes number by manually counting the co-localization events between mitochondrial (mtGFP) and autophagosomal marker (DsRed::LGG-1) displayed in each stack of body wall muscle cell (Figure 3).
- Process images acquired from (a) method with ImageJ software to measure the average pixel intensity values and total area for each fluorescent image of transgenic worm. Focus on body wall muscle cells of the head region avoiding intestinal autofluorescence (Figures 1 and 2; see Note 5). To analyze the area of interest manually:
Data analysis
- For each strain or condition, use at least 100 animals or 50 body wall muscle cells to obtain more accurate results.
- Each assay should be repeated at least three (3) times.
- Use the Student’s t-test with a significance cut-off level of P < 0.05 for comparisons between two groups.
- Use the one-factor (ANOVA) variance analysis and correct by the post hoc Bonferroni test for multiple comparisons.
Notes
- Transfer the selected transgenic animals to freshly seeded NGM plates every two days to avoid progeny and prevent starvation due to lack of food. Calorie deprivation and starvation induce mitophagy. Thus, well-fed animals should be used.
- The use of M9 buffer instead of water ensures a favourable osmotic environment for the nematodes.
- Levamisole is a mild anaesthetics and is required to immobilize nematodes. Avoid anaesthetics that could interfere with metabolic processes, such as sodium azide. Sodium azide induces mitochondrial and oxidative stress through inhibition of mitochondrial respiratory chain and perturbation of energy production. Thus, sodium azide is likely to induce mitophagy.
- Take a toothpick and glue an eyebrow/eyelash hair to the tip of it. Let it dry at room temperature. Then, use this tool to pick nematodes. Before using the eyebrow/eyelash hair always sterilize it by using 70% of EtOH.
- Intestinal autofluorescence increases during ageing in C. elegans. Thus, body wall muscle cells close to the intestine should be avoided during the imaging process.
Recipes
- Phosphate buffer (1 M)
- For 1 L, dissolve 102.2 g KH2PO4 and 57.06 g K2HPO4 in distilled water and fill up to 1 L. This is a 1 M solution, pH 6.0
- Autoclave and keep at room temperature
- Nematode growth medium (NGM) agar plates
- Mix 3 g NaCl, 2.5 g Bacto peptone, 0.2 g streptomycin, 17 g agar and add 900 ml distilled water. Autoclave
- Let cool to 55-60 °C
- Add 1 ml cholesterol stock solution, 1 ml 1 M CaCl2, 1 ml 1 M MgSO4, 1 ml nystatin stock solution, 25 ml sterile 1 M phosphate buffer, pH 6.0, and distilled sterile water up to 1 L
- Pour about 8 ml medium per Petri dish and leave to solidify
- Keep the plates at 4 °C until used
- M9 buffer
- Dissolve 3 g KH2PO4, 6 g Na2HPO4, 5 g NaCl in 1 L distilled water. Autoclave
- Let cool and add 1 ml 1 M MgSO4 (sterile)
- Store M9 buffer at 4 °C
- Levamisole (0.5 M)
- Dissolve 1.2 g levamisole in 10 ml distilled water
- Store levamisole solution at 4 °C
- M9-levamisole (20 mM)
- Dilute 400 μl 0.5 M levamisole in 10 ml M9 buffer
- Store M9-levamisole solution at 4 °C
- Paraquat (0.5 M)
- Dissolve 1 g paraquat in 8 ml distilled water
- Prepare aliquots of 400 ml and store them at 4 °C
- Carbonyl cyanide m-chlorophenylhydrazone (49 mM; CCCP)
- Dissolve 100 mg CCCP in 10 ml of DMSO
- Prepare aliquots of 1 ml and store them at -20 °C
Acknowledgments
This work was funded by grants from the European Research Council (ERC), the European Commission 7th Framework Programme and Bodossaki Foundation Postdoctoral Research Fellowship. The protocol has been adapted from Palikaras et al. (2015), Nature 521, 525-528.
References
- Fang, E. F., Kassahun, H., Croteau, D. L., Scheibye-Knudsen, M., Marosi, K., Lu, H., Shamanna, R. A., Kalyanasundaram, S., Bollineni, R. C., Wilson, M. A., Iser, W. B., Wollman, B. N., Morevati, M., Li, J., Kerr, J. S., Lu, Q., Waltz, T. B., Tian, J., Sinclair, D. A., Mattson, M. P., Nilsen, H. and Bohr, V. A. (2016). NAD+ replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab 24(4): 566-581.
- Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. and Kroemer, G. (2013). The hallmarks of aging. Cell 153(6): 1194-1217.
- Narendra, D. P., Jin, S. M., Tanaka, A., Suen, D. F., Gautier, C. A., Shen, J., Cookson, M. R. and Youle, R. J. (2010). PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 8: e1000298.
- Palikaras, K., Lionaki, E. and Tavernarakis, N. (2015). Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521(7553): 525-528.
- Palikaras, K. and Tavernarakis, N. (2014). Mitochondrial homeostasis: the interplay between mitophagy and mitochondrial biogenesis. Exp Gerontol 56: 182-188.
- Pickrell, A. M. and Youle, R. J. (2015). The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron 85(2): 257-273.
- Rosado, C. J., Mijaljica, D., Hatzinisiriou, I., Prescott, M. and Devenish, R. J. (2008). Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy 4(2): 205-213.
- Scheibye-Knudsen, M., Fang, E. F., Croteau, D. L., Wilson, D. M., 3rd and Bohr, V. A. (2015). Protecting the mitochondrial powerhouse. Trends Cell Biol 25(3): 158-170.
- Schiavi, A., Maglioni, S., Palikaras, K., Shaik, A., Strappazzon, F., Brinkmann, V., Torgovnick, A., Castelein, N., De Henau, S., Braeckman, B. P., Cecconi, F., Tavernarakis, N. and Ventura, N. (2015). Iron-starvation-induced mitophagy mediates lifespan extension upon mitochondrial stress in C. elegans. Curr Biol 25(14): 1810-1822.
- Vafai, S. B. and Mootha, V. K. (2012). Mitochondrial disorders as windows into an ancient organelle. Nature 491(7424): 374-383.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Palikaras, K. and Tavernarakis, N. (2017). In vivo Mitophagy Monitoring in Caenorhabditis elegans to Determine Mitochondrial Homeostasis. Bio-protocol 7(7): e2215. DOI: 10.21769/BioProtoc.2215.
Category
Developmental Biology > Cell signaling > Energy homeostasis
Developmental Biology > Cell signaling > Mitophagy
Cell Biology > Cell imaging > Live-cell imaging
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











