- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In Gel Kinase Assay
Published: Vol 7, Iss 5, Mar 5, 2017 DOI: 10.21769/BioProtoc.2170 Views: 16122
Reviewed by: Rainer MelzerHarrie van ErpAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1465 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3196 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1811 Views
Abstract
Proper spatiotemporal regulation of protein phosphorylation in cells and tissues is required for normal development and homeostasis. We present the protocol ‘In Gel Kinase Assay’, which is useful for protein kinase activity measurements from crude protein extracts. We have successfully used ‘In Gel Kinase Assay’ protocol to show that the Arabidopsis thaliana sextuple mutant in the PYRABACTIN RESISTANCE1/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS (PYR/PYL/RCAR-ABA receptors; line pyr/pyl112458) is impaired in ABA-mediated activation of SnRK2.2, SnRK2.3 and OST1/SnRK2.6, as much as the triple mutant snrk2.2/2.3/2.6 (Gonzalez-Guzman et al., 2012).
Keywords: KinasesBackground
The phytohormone abscisic acid (ABA) is a key signal involved in plant growth and development as well as in plant response to abiotic and biotic stress. The ABA perception and signaling pathway is composed of PYRABACTIN RESISTANCE1/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS (PYR/PYL/RCAR-ABA receptors), PP2Cs phosphatases and SnRK2s kinases (reviewed in Antoni et al., 2011). The module receptor-ABA-phosphatase controls the phosphorylation signaling cascades in a ligand-dependent manner through regulation of ABA-activated SnRK2s. In turn, SnRK2s kinases phosphorylate a myriad of effectors, both in the nucleus and in the cytoplasm, from transcription factors (e.g., ABFs) to ion channels (e.g., SLAC1). We show here the protocol ‘In Gel Kinase Assay’ in details. This protocol was developed for protein kinase activity measurement in plant tissues protein extracts as well as from purified recombinant kinases. In brief: an SDS-polyacrylamide gel is prepared containing the ∆C-ABF2 peptide (a specific SnRK2.2, SnRK2.3, OST1/SnRK2.6 kinase substrate). ∆C-ABF2 peptide is trapped in the SDS-polyacrylamide gel mesh and it does not migrate during electrophoresis. On the other hand, SDS-treated protein samples, without boiling and without any reducing agent (e.g., DTT or B-ME), are only partially denatured and can be re-naturalized in some degree after washing SDS out. Moreover, kinase activity at a high sensitive level can be measured using [Gamma-32p]ATP. In this way, together with the kinase activity value we also have the electrophoretic mobility value associated with the kinase activity. Using a plant crude protein extract, and after sample electrophoresis and in gel protein renaturalization step, we can measure specific SnRK2.2/2.3/2.6 (SnRK2s) kinase activity in gel. We have applied this protocol to characterize SnRK2s activity in a PYR/PYL/RCAR-ABA receptor sextuple mutant. We show that the sextuple mutant line pyr/pyl112458 is impaired in ABA-mediated activation of SnRK2s, as much as the triple mutant snrk2.2/2.3/2.6 in Arabidopsis thaliana (result published in Gonzalez-Guzman et al., 2012).
Materials and Reagents
- Eppendorf and Falcon tubes (1.5 ml and 50 ml respectively; generic)
- Pipettes and tips (generic)
- Micropore tape (3M, catalog number: 1530-0 )
- Sterile round Petri dishes (100 mm diameter x 20 mm height; generic: e.g., Greiner Bio One, catalog number: 664161 )
- Extra thick blot filter paper, 7.5 x 10 cm (Bio-Rad Laboratories, catalog number: 1703965 )
- Arabidopsis thaliana seeds: Col-0 (NASC, N1093), pyr/pyl112458 and snrk2.2/2.3/2.6 (Gonzalez-Guzman et al., 2012)
- Optional: purified recombinant kinase OST1 (SnRK2.6-6his; homemade: Vlad et al., 2009 and 2012)
- Sterile deionized water (generic)
- Liquid N2 (generic)
- Ice (generic)
- ABA (+cis, trans-Abscisic acid) (BIOSYNTH, catalog number: A-0120 )
- Bio-Rad Protein Assay Kit I (Bio-Rad Laboratories, catalog number: 5000001 )
- Bio-SafeTM Coomassie Stain reagent (Bio-Rad Laboratories, catalog number: 1610786 )
- ∆C-ABF2 peptide (amino acids 1 to 173; 18.4 KD; homemade: Dupeux et al., 2011; Antoni et al., 2012)
Note: Commercially available alternative MBP (myelin basic protein) (Sigma-Aldrich, catalog number: M1891 ). - SDS-PAGE reagents (listed in He, 2011)
- Ethanol (generic)
- Polyethylene glycol sorbitan monolaurate (Tween-20) (Sigma-Aldrich, catalog number: P1379 )
- Murashige and Skoog medium (MS) (PhytoTechnology Laboratories®, catalog number: M524 )
- 2-N-morpholino-ethane-sulfonic acid (MES) (Sigma-Aldrich, catalog number: M8250 )
- Sucrose (VWR, BDH®, catalog number: BDH0308 )
- Agar-agar (Sigma-Aldrich, catalog number: A1296 )
- Potassium hydroxide (KOH) (Fisher Scientific, catalog number: P250 )
- 2-Amino-2-(hydroxymethyl)-1,3-propanediol (Tris-base) (Sigma-Aldrich, catalog number: 252859 )
- HCl
- 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid, N-(2-hydroxyethyl)piperazine-N’-(2-ethanesulfonic acid) (HEPES) (Sigma-Aldrich, catalog number: H3375 )
- DL-dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43815 )
- EDTA (Sigma-Aldrich, catalog number: E5134 )
- EGTA (Sigma-Aldrich, catalog number: E3889 )
- Sodium fluoride (NaF) (Sigma-Aldrich, catalog number: S7920 )
- Sodium orthovanadate (Na3VO4) (Sigma-Aldrich, catalog number: 450243 )
- Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- Adenosine 5’-triphosphate disodium salt hydrate (ATP) (Sigma-Aldrich, catalog number: A1852 )
- Glycerol 2-phosphate disodium salt hydrate (BGP) (Sigma-Aldrich, catalog number: G9422 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: S8830 )
- Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L3771 )
- 2-(5-Bromo-2-pyridylazo)-5-(diethylamino) (Bromo-phenol-blue) (Sigma-Aldrich, catalog number: 180017 )
- Polyethylene glycol tert-octylphenyl ether (Triton X-100) (Sigma-Aldrich, catalog number: T8787 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: 05470 )
- [Gamma-32p]ATP, 10 μCi/ml - 3,000 Ci/mmol (PerkinElmer, catalog number: BLU502A )
- Trichloroacetic acid (TCA) (Sigma-Aldrich, catalog number: T6399 )
- Sodium pyrophosphate (Na2PPi) (Sigma-Aldrich, catalog number: 71501 )
- Seed sterilization solution (see Recipes)
- Solid MS (see Recipes)
- Stock reagent (see Recipes)
- 1 M Tris-HCl (pH 6.8 and 7.5)
- 1 M HEPES-KOH (pH 7.5)
- 1 M DTT
- 0.5 M EGTA
- 0.5 M EDTA
- 1 M NaF
- 100 mM Na3VO4
- 100 mM PMSF
- 1 M MgCl2
- 100 mM ATP
- Protein extraction buffer (see Recipes)
- 2x Laemli buffer (see Recipes)
- Washing buffer (see Recipes)
- Protein renaturalization buffer (see Recipes)
- Cold reaction kinase buffer (see Recipes)
- Hot reaction kinase buffer (see Recipes)
- STOP-washing buffer (see Recipes)
Equipment
- Radiation safety gears and personal protection (generic)
- Pipette
- Flow hood (generic: e.g., CLEATECH, catalog number: 1000-11-E )
- Plant growth chamber (generic: e.g., BioChambers, model: FXC-9 )
- Benchtop microcentrifuge (generic: e.g., Thermo Fisher Scientific, Thermo ScientificTM, model: SorvallTM LegendTM Micro 17 , catalog number: 75002430)
- SDS-PAGE system (Bio-Rad Laboratories, model: Mini-PROTEAN® Tetra Vertical Electrophoresis Cell )
- Vacuum gel dryer (generic: e.g., Bio-Rad Laboratories, model: 583 Gel Dryers )
- Phosphorimager screen and cassette (FUJIFILM)
- Phosphorimager (FUJIFILM, model: FLA-5100 )
- Autoclave (generic)
- Mortar (100 mm diameter x 60 mm height; generic)
- Spatula and forceps (generic)
- Plastic incubation box (for Polyacrylamide gel incubation and washing; generic)
- Stirring bars (50 mm long; generic)
- Timer (generic)
- Rotary shaker (generic)
- Fridge (4 °C; generic)
- Freezer (-20 °C and -80 °C; generic)
- Magnetic stirrer (generic)
- pH meter (generic: e.g., Thermo Fisher Scientific, Thermo ScientificTM, model: Orion StarTM A111 )
- Balance (generic: e.g., Sartorius, model: Cubis® Precision Balance )
- Vortex (generic)
- Spectrophotometer (generic: e.g., Thermo Fisher Scientific, Thermo ScientificTM, model: EvolutionTM 300 )
Software
- ImageJ (optional)
Procedure
Note: Although this protocol was developed for plant tissue protein extracts as well as recombinant proteins, it potentially can also be applied for protein extracts from any other source. Even more, this protocol could also be valid for samples obtained from immunoprecipitation assay (Vlad et al., 2009).
- Seed sterilization and plant growth
- Put 5 mg (200-300 seeds) of Arabidopsis seeds into an Eppendorf tube, add 750 μl sterilization solution (Recipe 1) and shake for 20 min at room temperature.
Note: In this protocol we tested: pyr/pyl sextuple mutant line (pyr/pyl112458), snrk triple mutant line (snrk2.2/2.3/2.6) and Col-0 as a control line (described in Gonzalez-Guzman et al., 2012). - Remove the sterilization solution (by pipetting with sterile tips) in flow hood and wash seeds with 750 μl sterile distilled water (shaking it for 15 min at room temperature). Repeat this step 3 times.
- Stratify seeds in 750 μl fresh sterile distilled water at 4 °C for 3 days in the dark.
- Sow seeds on sterile solid MS plates (Recipe 2).
Note: Keep a density of approximately one seed per cm2 (crowded plates produce unhealthy seedlings). - Close and seal plates with micropore tape.
Note: Use only one layer of micropore tape. More than one layer prevents both: gaseous exchange and healthy plant growth. - Incubate plates for 12 days in a controlled environment growth chamber at 22 °C under a 16-h light/8-h dark photoperiod at 100 µEm-2 sec-1.
Note: Time and growth condition could be variable depending on experimental requirements.
- Put 5 mg (200-300 seeds) of Arabidopsis seeds into an Eppendorf tube, add 750 μl sterilization solution (Recipe 1) and shake for 20 min at room temperature.
- Total protein extraction and SDS-PAGE
- Harvest seedlings from plates (500 mg) and freeze it in liquid N2. Store at -80 °C until use.
Note: In our experimental design 100 µM ABA or mock treatment (sprayed - 30 min) was required before freezing. - Grind plant tissues into fine powder using mortar and liquid N2.
- Transfer 400 mg of the fine powder into a 2 ml Eppendorf tube using a cooled (N2) spatula; add 1 ml of cold extraction buffer (Recipe 3) and mix well by vortexing for 15 sec.
Note: Keep tube on ice until centrifugation (proceed with the next sample). - Centrifuge for 15 min at 4 °C (maximum speed, 14,000 x g).
- Transfer supernatant into a new 1.5 ml Eppendorf tube and store at -80 °C until use.
Note: Before storing tubes at -80 °C, take a sample of each one (50 µl) for protein quantification. Repeated sample freeze and thaw can decrease enzymatic activity. - Proceed with protein quantification.
Note: Any protein quantification method should be fine. We choose Bradford Protein Assay from Bio-Rad (follow manufacturers’ protocol) - Prepare 10% polyacrylamide gel (for SDS-PAGE) and include before polymerization 800 µg of ∆C-ABF2 peptide (in the resolving gel).
Notes: - We choose miniprotean III SDS-PAGE system from Bio-Rad and a gel of 1.5 mm width.
- We use as specific SnRK2.2/2.3/.2.6 kinase substrate the His6-∆C-ABF2 peptide (amino acids 1 to 173; 18.4 KD). ∆C-ABF2 cloning and recombinant protein preparation was described in: Dupeux et al., 2011 and Antoni et al., 2012 (see a brief description of these methods in Notes section).
- Potentially other recombinant kinase substrates could be used depending on which kinases will be assayed. Even synthetic or commercially available peptides, i.e., MBP (myelin basic protein), should be suitable.
- For SDS-PAGE preparation follow the BIOPROTOCOL e80 (He, 2011)
- Prepare loading samples: mix 40 µg of total protein plant extract (obtained in step B5 and thawed on ice) with 50 µl 2x Laemli buffer (Recipe 5) and add sterile deionized H2O to reach 100 µl final volume.
Notes: - Do not boil the samples. Boiling produces irreversible protein denaturation and loss of enzymatic activity. (Important)
- Quantity of total protein extract could be variable depending on kinase expression and activity. For a pilot experiment, we suggest a concentration range from 20 µg to 80 µg of total protein extract.
- For recombinant protein study, active His-tagged OST1 protein kinase produced in E. coli was used (autophosphorylated OST1/SnRK2.6-6HIS, Vlad et al., 2009; see Notes section for recombinant protein purification). Different recombinant OST1/SnRK2.6-6HIS amounts where tested: 0.01, 0.05, 0.1, 0.5, 1, 5 and 10 µg.
- Incubate loading samples on ice for 15 min.
- Load and run SDS-PAGE over-night (10-12 h) at constant 40 V.
Notes: - Optionally run SDS-PAGE for 4-5 h at constant 80 V.
- Run a parallel SDS-PAGE (without ∆C-ABF2 peptide) and load the same sample amount. After running, stain this gel with Coomassie to check protein loaded into gel. This Coomassie stained gel is also useful for densitometric analysis if required.
- Harvest seedlings from plates (500 mg) and freeze it in liquid N2. Store at -80 °C until use.
- In-gel kinase activity reaction
Note: This protocol utilizes radioactive material and operators require experience in personal and environmental radiation protection and in the management of radioactive waste. (Important)- Gel washing: wash gel 3 times (30 min each) with 100 ml of washing buffer (Recipe 5) at room temperature and gentle agitation.
- Protein renaturalization: incubate gel 3 times (first 2 times 30 min each, and the last incubation over-night 10-12 h) with 150 ml of protein renaturalization buffer (Recipe 6) at room temperature. Gentle agitation is required.
- Pre-reaction incubation: incubate gel 30 min in 40 ml of cold reaction kinase buffer (Recipe 7), at room temperature and with gentle agitation.
- Kinase reaction activity: incubate gel 1 h in 8 ml of hot reaction kinase buffer (Recipe 8). Gentle agitation and room temperature are required.
Note: Prepare the minimum quantity possible of hot reaction kinase buffer to reduce radioactive waste. - Kinase STOP reaction: wash gel 6 times (1 h each) with STOP-washing buffer (Recipe 9). Gentle agitation and room temperature are required.
- Gel drying: dry gel over a filter paper with a gel vacuum dryer.
- Radioactivity detection: detect radioactivity using screen and cassette (FUJIFILM) with variable exposure times (from 1 to 72 h) and a phosphorimage detector system (FLA5100; FUJIFILM).
- Gel washing: wash gel 3 times (30 min each) with 100 ml of washing buffer (Recipe 5) at room temperature and gentle agitation.
Data analysis
In Figure 1 we show activity of His-tagged OST1 protein kinase (OST1/SnRK2.6-6HIS; Vlad et al., 2009) produced in E. coli. This kinase became autophosphorylated and active in absence of PP2Cs phosphatases. As expected, we see an increment in OST1 activity when we load more amount of the purified protein sample. On the other hand, we monitored the in vivo activation status of SnRK2s using crude protein extracts from wild-type (Col-0) and mutant lines: pyr/pyl112458 and snrk2.2/2.3/2.6 (Figure 2). The ‘In Gel Kinase Assay’ show kinase activity in the three ABA-activated SnRK2s (SnRK2.2/2.3/2.6), identified as a double band between 42 to 44 kD, in ABA-treated (100 µM ABA) Col-0. By contrast, as expected, this SnRK2s kinase activity was absent in control conditions Col-0 (mock treated) and in the ABA-treated (100 µM ABA) mutant line snrk2.2/2.3/2.6. Interestingly, the ‘In Gel Kinase Assay’ did not detect any activation of the SnRK2s in the sextuple mutant line pyr/pyl112458 after 100 µM ABA treatment. This result supports the idea that PYR/PYL112458 receptors are the main players in ABA perception and signaling.
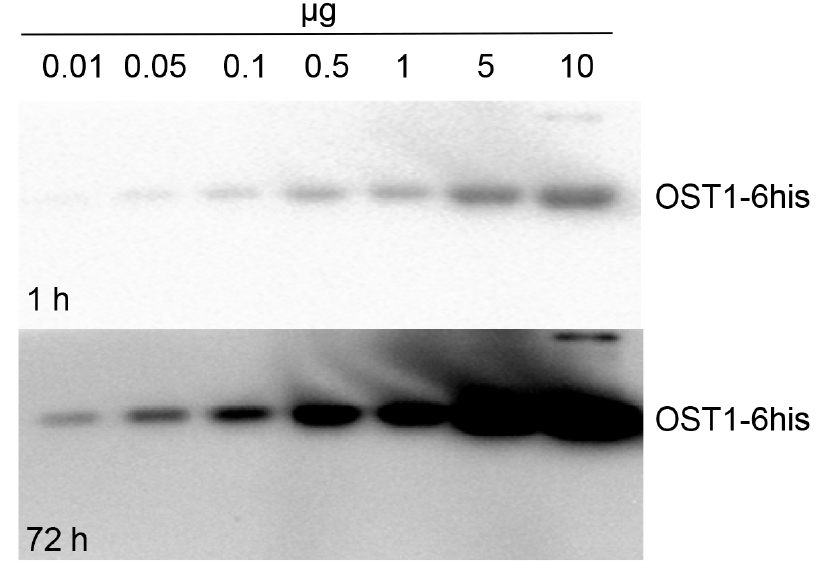
Figure 1. Kinase activity measurement of recombinant OST1/SnRK2.6-6HIS. OST1/SnRK2.6-6HIS was produced in E. coli and purified by Ni affinity. Different recombinant protein amount where tested: 0.01, 0.05, 0.1, 0.5, 1, 5 and 10 µg. 1 h and 72 h: exposure time.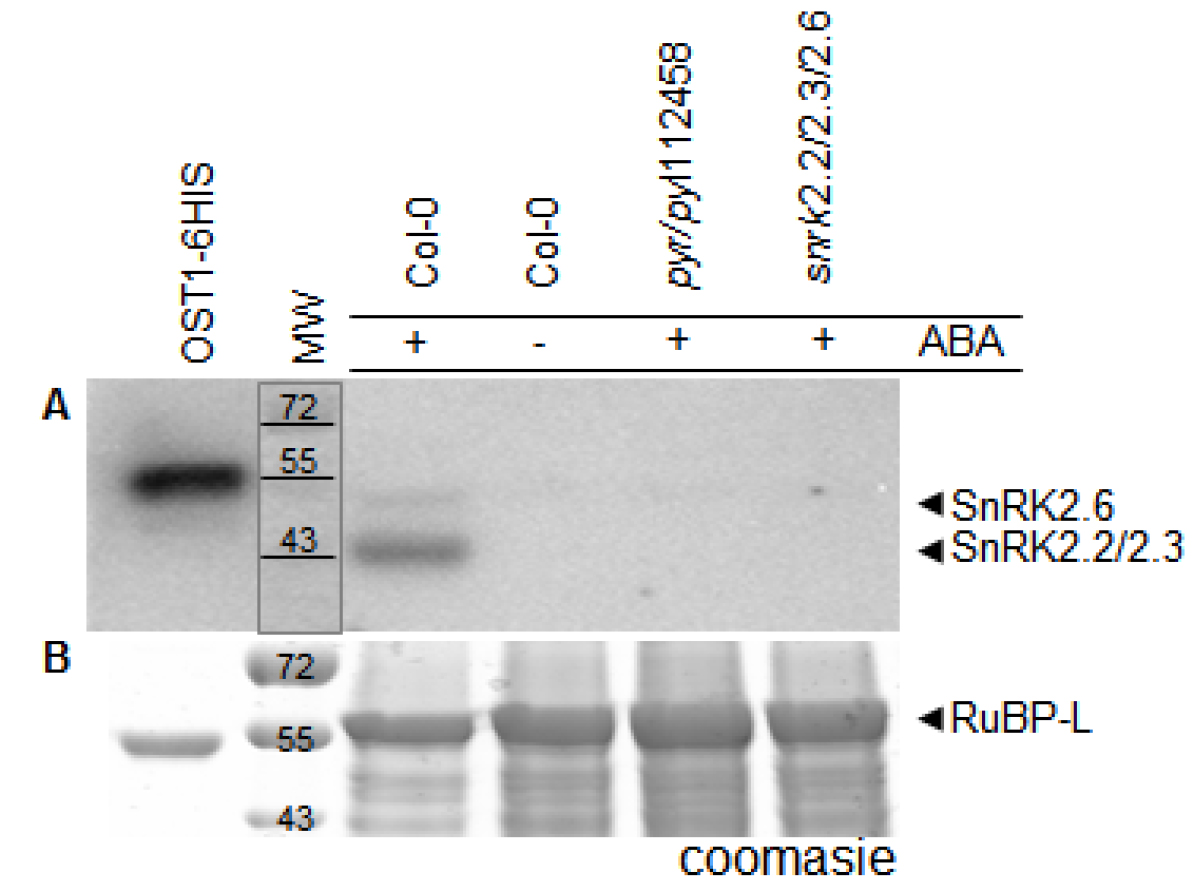
Figure 2. In vivo monitoring of SnRK2s activity. Wild-type (Col-0) and mutant lines: pyr/pyl112458 and snrk2.2/2.3/2.6 were mock or ABA (100 µM) treated for 30 min. A. SnRK2.2/2.3/2.6 kinase activity is detectable in ABA-treated (100 µM ABA) Col-0 and it is not detected in Col-0 control conditions (mock treated) neither ABA-treated (100 µM ABA) mutant line snrk2.2/2.3/2.6 nor sextuple mutant line pyr/pyl112458. Recombinant OST1/SnRK2.6-6HIS was used as a positive control. B. A parallel Coomassie stained SDS-PAGE. MW: molecular weight [kD]: 72, 55 and 43. RuBP-L: RuBisCO large chain.
Note: Given the necessity, substrate phosphorylation by SnRK2.2/2.3/2.6 over time can be quantified by densitometric analysis using open source software (e.g., ImageJ). Band signal volume (band area x band total intensity) in the phosphorylation assay gel must be calculated and the protein loading in the SDS-PAGE should be checked (a parallel Coomassie stained SDS-PAGE is an option).
Notes
- This technique does not present major problems in its execution, beyond those related to the radioactive ATP handling. To make a parallelism, the success in this technique may be similar to that obtained in Western-blots. In particular, the success rate of ‘In Gel Kinase Assay’ measuring SnRK2.2/2.3/2.6 kinase activity may be similar to the Western-blot technique using a good primary antibody.
- The triple mutant snrk2.2/2.3/2.6 was used as a negative control (no ABA-induced SnRK2s kinase activity). For snrk2.2, snrk2.3, snrk2.6 single mutant, snrk2.2/2.3 double mutant and snrk2.2/2.3/2.6 triple mutant characterization read Fujii et al. (2007 and 2009).
- Brief protocol for small-scale recombinant protein purification (from Antoni et al., 2012):
- E. coli BL21 (DE3) cells transformed with the corresponding constructs (OST1/SnRK2.6-6HIS and ∆C-ABF2, both in pET28a E. coli expression plasmid) were grown in 100 ml of LB medium to an optical density at 600 nm of 0.6 to 0.8.
- At this point, 1 mM isopropyl-b-D-thiogalactoside (IPTG) was added, and the cells were harvested after overnight incubation at 20 °C.
- Pellets were resuspended in lysis buffer (50 mM Tris, pH 7.5, 250 mM KCl, 10% [v/v] glycerol, and 1 mM b-mercaptoethanol) and lysed by sonication with a Branson Sonifier 250.
- The clear lysate obtained after centrifugation was purified by Ni affinity.
- A washing step was performed using 50 mM Tris, 250 mM KCl, 20% (v/v) glycerol, 30 mM imidazole, and 1 mM b-mercaptoethanol washing buffer, and finally the protein was eluted using 50 mM Tris, 250 mM KCl, 20% (v/v) glycerol, 250 mM imidazole, and 1 mM b-mercaptoethanol elution buffer.
For an extended protocol of ‘Recombinant Protein Expression and Purification’ see this Bio-Rad protocol: http://www.bio-rad.com/webroot/web/pdf/lse/literature/pepsi_hr_1665067.pdf
Recipes
- Seed sterilization solution
70% (v/v) ethanol
0.05% (v/v) Tween-20 - Solid MS
4.33 g/L Murashige & Skoog basal salts
0.1% (w/v) MES
1% (w/v) sucrose
1% (w/v) agar
Adjust pH to 5.7 with KOH before autoclaving - Stock reagent
- 1 M Tris-HCl (pH 6.8 and 7.5), for 50 ml: 6.05 g Tris-base + 45 ml ddH2O. Bring to pH 6.8 or 7.5 with HCl and adjust volume with ddH2O to 50 ml (store at room temperature)
- 1 M HEPES-KOH (pH 7.5), for 50 ml: 14.2 g HEPES + 45 ml ddH2O. Bring to pH 7.5 with KOH and adjust volume with ddH2O to 50 ml (store at room temperature)
- 1 M DTT, for 50 ml: 7.5 g DTT in 50 ml ddH2O (store in aliquots at -20 °C)
- 0.5 M EGTA, for 50 ml: 9.5 g EGTA + 45 ml ddH2O. Bring to pH 8 with NaOH and adjust volume with ddH2O to 50 ml (store at room temperature)
- 0.5 M EDTA, for 50 ml: 9.3 g EDTA + 45 ml ddH2O. Bring to pH 8 with NaOH and adjust volume with ddH2O to 50 ml (store at room temperature)
- 1 M NaF, for 50 ml: 2.1 g NaF and adjust volume with ddH2O to 50 ml (store at 4 °C)
- 100 mM Na3VO4, for 100 ml: 1.82 g Na3VO4 + 90 ml ddH2O. Bring to pH 9 with HCl and boil solution until colorless. Cold solution to room temperature, adjust again to pH 9 with HCl and boil solution again (repeat this process until pH remain stable at pH 9 at room temperature). Adjust volume with ddH2O to 50 ml (store at -20 °C)
- 100 mM PMSF, for 10 ml: 174 mg PMSF adjust volume with ethanol to 10 ml (store in aliquots at -20 °C)
- 1 M MgCl2, for 10 ml: 4.76 g MgCl2 adjust volume with ddH2O to 10 ml (store in aliquots at -20 °C)
- 100 mM ATP, for 10 ml: 551 mg ATP adjust volume with ddH2O to 10 ml (store in aliquots at -20 °C)
- Protein extraction buffer
5 mM EDTA
5 mM EGTA
2 mM DTT
25 mM NaF
1 mM Na3VO4
50 mM BGP
20% (v/v) glycerol
1 mM PMSF
1 tablet/100 ml protease inhibitor cocktail
50 mM HEPES-KOH (pH 7.5) - 2x Laemli buffer
125 mM Tris-HCl, pH 6.8
4% (w/v) SDS
20% (v/v) glycerol
0.001% bromo-phenol-blue - Washing buffer
25 mM Tris-HCl (pH 7.5)
0.1 % (v/v) Triton X-100
0.05 % (w/v) BSA
0.5 mM DTT
0.1 mM Na3VO4
5 mM NaF - Protein renaturalization buffer
25 mM Tris-HCl (pH 7.5)
1 mM DTT
0.1 mM Na3VO4
5 m M NaF - Cold reaction kinase buffer
40 mM HEPES-KOH (pH 7.5)
1 mM EGTA
200 mM MgCl2
2 mM DTT
0.1 mM Na3VO4 - Hot reaction kinase buffer
8 ml cold reaction kinase buffer
25 µM cold ATP (no radioactive)
80 µCi hot ATP (radioactive) - STOP-washing buffer
5% (w/v) TCA (trichloroacetic acid)
1% (p/v) Na2PPi (sodium pyrophosphate)
Acknowledgments
We thank Prof. Christiane Laurière for giving us kind assistance in the development of this protocol. This work was supported by the Ministerio de Ciencia e Innovación, Fondo Europeo de Desarrollo Regional, and Consejo Superior de Investigaciones Científicas (grant BIO2014-52537-R to P.L.R). This protocol was adapted from the research article of Gonzalez-Guzman et al. (2012). The author declares no conflicts of interest.
References
- Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., Rodrigues, A., Pizzio, G. A. and Rodriguez, P. L. (2012). Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiol 158(2): 970-980.
- Antoni, R., Rodriguez, L., Gonzalez-Guzman, M., Pizzio, G. A. and Rodriguez, P. L. (2011). News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Curr Opin Plant Biol 14(5): 547-553.
- Dupeux, F., Antoni, R., Betz, K., Santiago, J., Gonzalez-Guzman, M., Rodriguez, L., Rubio, S., Park, S. Y., Cutler, S. R., Rodriguez, P. L. and Marquez, J. A. (2011). Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiol 156(1): 106-116.
- Fujii, H., Verslues, P. E. and Zhu, J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19(2): 485-494.
- Fujii, H. and Zhu, J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci U S A 106(20): 8380-8385.
- Gonzalez-Guzman, M., Pizzio, G. A., Antoni, R., Vera-Sirera, F., Merilo, E., Bassel, G. W., Fernandez, M. A., Holdsworth, M. J., Perez-Amador, M. A., Kollist, H. and Rodriguez, P. L. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24(6): 2483-2496.
- He, F. (2011). Laemmli-SDS-PAGE. Bio-protocol Bio101: e80.
- Vlad, F., Rubio, S., Rodrigues, A., Sirichandra, C., Belin, C., Robert, N., Leung, J., Rodriguez, P. L., Lauriere, C. and Merlot, S. (2009). Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell 21(10): 3170-3184.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Pizzio, G. A. and Rodruiguez, P. L. (2017). In Gel Kinase Assay. Bio-protocol 7(5): e2170. DOI: 10.21769/BioProtoc.2170.
- Gonzalez-Guzman, M., Pizzio, G. A., Antoni, R., Vera-Sirera, F., Merilo, E., Bassel, G. W., Fernandez, M. A., Holdsworth, M. J., Perez-Amador, M. A., Kollist, H. and Rodriguez, P. L. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24(6): 2483-2496.
Category
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Biochemistry > Protein > Electrophoresis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











