- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Expression and Purification of Cyanobacterial Circadian Clock Protein KaiC and Determination of Its Auto-phosphatase Activity
Published: Vol 7, Iss 4, Feb 20, 2017 DOI: 10.21769/BioProtoc.2140 Views: 8118
Reviewed by: Dennis NürnbergAnna A. ZorinaEsteban Paredes-Osses

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Monitoring Protein Stability In Vivo Using an Intein-Based Biosensor
John S. Smetana [...] Christopher W. Lennon
Apr 20, 2025 1575 Views

Endo-1,4-β-D-xylanase Assay Using Azo-Xylan and Variants Thereof
Luca Bombardi [...] Salvatore Fusco
Apr 20, 2025 1916 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2167 Views
Abstract
Circadian rhythms are biological processes displaying an endogenous oscillation with a period of ~24 h. They allow organisms to anticipate and get prepared for the environmental changes caused mainly by the rotation of Earth. Circadian rhythms are driven by circadian clocks that consist of proteins, DNA, and/or RNA. Circadian clocks of cyanobacteria are the simplest and one of the best studied models. They contain the three clock proteins KaiA, KaiB, and KaiC which can be used for in vitro reconstitution experiments and determination of the auto-phosphatase activity of KaiC as described in this protocol.
Keywords: KaiABackground
The rotation of planet Earth causes the ~24 h day-night oscillation. To fit in and then efficiently take advantage of this rhythmic change of the environment, most if not all organisms have an endogenous activity rhythm of ~24 h, which is called circadian rhythm. Circadian rhythms provide evolutionary advantages to those organisms. The long-term disruption of circadian rhythms is extremely harmful (Ma et al., 2013). In humans, many diseases, including cancer, hypertension, and sleep disorders, are closely related with a disrupted circadian rhythm (Shi et al., 2013; Roenneberg and Merrow, 2016).
Circadian rhythms are controlled by endogenous rhythm generators called circadian clocks. A functional circadian clock has three functionalities: accepting the environmental information, turning the environmental cues into oscillating signals, and relaying these signals to down-stream modulators (Pattanayak and Rust, 2014). Cyanobacteria are the simplest organisms having a well-studied circadian clock, in which the oscillation generator is controlled by three proteins: KaiA, KaiB, and KaiC (Mackey et al., 2011; Johnson et al., 2011; Chen et al., 2013; Egli and Johnson, 2013). KaiC is a multi-functional protein, which has auto-kinase, auto-phosphatase, and ATPase activity (Egli, 2015). The auto-kinase activity results in the phosphorylation of the two key residues T432 and S431 in KaiC, whereas the auto-phosphatase activity results in their de-phosphorylation (Rust, 2012). When incubated alone, KaiC shows mainly phosphatase activity (Nishiwaki and Kondo, 2012). KaiA can stimulate the auto-kinase activity of KaiC, and KaiB antagonizes KaiA’s function, which makes the phosphorylation state of KaiC oscillate in a ~24 h rhythm (Dong et al., 2016).
In 2005, Nakajima et al. successfully reconstituted the KaiABC oscillator in vitro by mixing the purified proteins, KaiA, KaiB, and KaiC, in a buffer containing ATP and Mg2+ (Nakajima et al., 2005). The simple procedure made the KaiABC system a highly attractive model for studying the molecular mechanism of circadian clocks. In this protocol, a major part of the reconstitution system, the in vitro determination of the auto-phosphatase activity of KaiC is described, in which the phosphorylation states of KaiC are analyzed by SDS-PAGE.
Materials and Reagents
- 1.5 ml microcentrifuge tubes
- 15 ml tube (Corning, Axygen®, catalog number: SCT-15mL-25-S )
- 50 ml centrifuge tube (Corning, Axygen®, catalog number: SCT-50mL-25-S )
- Petri dish (Corning, catalog number: 70165-60 )
- Hitrap FF Q column: 5 ml (GE Healthcare, catalog number: 17-5156-01 )
- NAP-5/25 buffer exchange column (GE Healthcare, catalog number: 17-0853-02 )
- Centrifugal filter: 10 kDa, Amicon Ultra (EMD Millipore, catalog number: PR02967 )
- 0.22 µm filter (Pall, catalog number: PN 4612 )
- PCR tube (Bio-Sharp, catalog number: BS-02-P )
- 0.45 µm filter
- E. coli strain BL21 (DE3) (New England Biolabs, catalog number: C2527 )
- pGEX-6P-1-KaiC: Provided by Prof. Carl Johnson (Vanderbilt University, USA). The KaiC-coding sequence is from Synechococcus elongatus PCC 7942
- Bacto-tryptone (Oxoid, catalog number: LP0042 )
- Bacto-yeast extract (Oxoid, catalog number: LP0021 )
- Agar A (Beijing Dingguo Changsheng Biotechnology, catalog number: DH010 )
- Calcium chloride (CaCl2) (1 M; Sigma-Aldrich, catalog number: V900266 )
- Ampicillin (1 mg/ml; North China Pharmaceutical Group Corporation, catalog number: A102048-25g )
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sigma-Aldrich, catalog number: 16758 )
- Glutathione S-transferase (GST) resin (EMD Millipore, catalog number: 70541 )
- PreScission protease (PSP) (GE Healthcare, catalog number: 27-0843-01 )
- Kanamycin (1 mg/ml; GENVIEW, catalog number: AK177-10G )
- 3x loading dye
- Tris-base (AMRESCO, catalog number: 0497 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: 7647-14-5 )
- Dithiothreitol (DTT) (Life Science Products&Services, catalog number: DB0058-25g )
- Tween-20 (Enox, catalog number: 557 )
- ATP (Life Science Products&Services, catalog number: AB0020-25g )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- EDTA (Bio Basic, catalog number: EB0185 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- SDS (Life Science Products&Services, catalog number: SB0485 )
- Bis-acrylamide (Sigma-Aldrich, catalog number: 146072 )
- Ammonium persulfate (APS) (Xilong Scientific, catalog number: 51504 )
- TEMED (CUSABIO, catalog number: V900853 )
- Pierce Coomassie (Bradford) Protein Assay Kit (Sangon Biotech, catalog number: C503031 )
- Bromophenol blue (Sigma-Aldrich, catalog number: 115-39-9 )
- Methanol (Tianjin Kermel Chemical Reagent, catalog number: 32058 )
- Acetic acid (Heng Xing, catalog number: 81601 )
- Acrylamide (AMRESCO, catalog number: 0341 )
- Protein marker (SM0431, Fermantas) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 26610 )
- GSH (Reduced Glutathione) (Sangon Biotech, catalog number: A100399 )
- Ethanol (Heng Xing, catalog number: 32061 )
- PSP buffer (see Recipes)
- Buffer A (see Recipes)
- Buffer B (see Recipes)
- Reaction buffer (see Recipes)
- 1 M IPTG (see Recipes)
- 10x running buffer (see Recipes)
- 8% separation gel (see Recipes)
- 5% stacking gel (see Recipes)
- Coomassie blue staining solution (see Recipes)
- De-staining buffer (see Recipes)
Equipment
- Autoclave sterilizer (Shanghai Huaxian Medical Equipment, model: HVA-85 )
- Laminar flow hood (SU ZHOU AN TAI, model: SW-CJ-2F )
- Electric Thermostatic incubator (Shanghai Yuejin Medical Apparatus Factory, model: SHP-250 )
- Pipettes (Gilson, catalog numbers: 711111170000 and 711111050000 )
- Orbital shaker (Thermo Fisher Scientific, Thermo ScientificTM, model: SHKE4000-1CE )
- High-speed refrigerated centrifuge (Hitachi, model: CR21G )
- Refrigerated centrifuge (Sigma Laborzentrifugen, model: 5811XQ 14241g )
- Benchtop centrifuge (Eppendorf, model: 1024 and 5424 )
- Ultra-low temperature freezer (Haier, model: 906 )
- Microplate spectrophotometer (Thermo Fisher Scientific, model: 1500 )
- Analytical balance (Sartorius, model: CP225D )
- Ultrapure water system (Pall, model: CascadA ZX )
- Membrane filter system (EMD Millipore, USA)
- Ultrasonic cell processor (Scientz Biotechnology, model: SCIENTZ-IID )
- FPLC system (GE Healthcare, model: AKTA Purifier 100 )
- Precision pH meter (Mettler Toledo, model: EL-20 )
- Electrophoresis system (Bio-Rad Laboratories, model: Mini-Protean Tetra )
- Decolorization shaker (Haimen Kylin-Bell Lab Instruments, model: TS-8 )
- Gel imaging system (Kodak, model: Gel Logic 200 )
- PCR system (Biometra, model: T-Gradient Thermoblock )
Software
- ImageJ (version 1.8.0_77, NIH, USA)
- Excel (version 2012, Microsoft, USA)
Procedure
- Transformation of E. coli cells with plasmid the pGEX-6P-1-KaiC
- Place the calcium competent BL21(DE3) cells and the pGEX-6P-1-KaiC plasmid in ice water.
- Measure the plasmid concentration spectrophotometrically and add 0.1 µg of pGEX-6P-1-KaiC into 100 µl of BL21(DE3) cells in a 1.5 ml tube.
- Mix the plasmid and the cells gently and set down in ice water for 30 min.
- Place the tube in a 42 °C water bath for 90 sec before placing it in ice water for 60 sec.
- Add 400 µl of LB (Luria-Bertani) medium (without antibiotics) to the tube, and let the cells recover in a shaker at 37 °C, 150 rpm for 45 min.
- Spin down the cells at 6,000 x g (30 sec).
- Discard supernatant and re-suspend cells in 100 µl of fresh LB medium (without antibiotics).
- Spread cells evenly on solid LB medium supplemented with 100 µg/ml of ampicillin.
- Incubate the plate at 37 °C overnight until colonies form.
- Place the calcium competent BL21(DE3) cells and the pGEX-6P-1-KaiC plasmid in ice water.
- Expression of KaiC
- Pick a single colony of the transformed BL21(DE3) cells containing pGEX-6P-1-KaiC and inoculate it into 3 ml liquid LB medium supplemented with 100 µg/ml of ampicillin in a sterilized 15 ml tube.
- Grow the cells overnight at 37 °C, 220 rpm.
- Transfer 1 ml of the overnight culture to 1 L of sterilized liquid LB medium containing 100 µg/ml of ampicillin in a 3 L flask.
- Grow culture until OD600 reaches ~0.6 at 37 °C, 220 rpm, lower the temperature to 28 °C and induce the expression of kaiC by adding 200 µl of 1 M IPTG.
- After 16 h of induction, collect the cells by centrifugation for 5 min at 7,000 x g, 4 °C.
- Keep the pelleted cells on ice for instant processing or store at -80 °C.
- Pick a single colony of the transformed BL21(DE3) cells containing pGEX-6P-1-KaiC and inoculate it into 3 ml liquid LB medium supplemented with 100 µg/ml of ampicillin in a sterilized 15 ml tube.
- GST affinity purification
- Re-suspend the cells from 1 L cell culture thoroughly with 40 ml of PSP buffer in a 100 ml glass beaker.
- Place the beaker in ice water, and disrupt the cells by sonication for 45 min at 35% output power (wait for 2 sec after every 1 sec of sonication).
- Transfer the solution to a fresh 50 ml tube, and centrifuge for 40 min at 4 °C, 16,000 x g.
- During the centrifugation, take 1 ml of the GST resin and wash it with 5 ml of sterilized ddH2O and then 3 ml of PSP buffer.
- Mix the GST resin on an orbital shaker with the supernatant from the centrifuge tube (step C3) for 2 h at 4 °C.
- Re-suspend the resin with 5 ml of PSP buffer (4 °C) and elute the buffer to remove unbound proteins.
- Add the PreScission protease to the resin and keep the solution at 4 °C for 16 h to allow the enzymatic digestion of the GST-tagged KaiC.
- Keep the supernatant (or the eluent) containing KaiC at 4 °C for further purification steps.
- Use an orbital shaker (40 rpm) when incubating the resin mixture at 4 °C.
- Store 10 µl of the sample at each step (including the steps in the following anion exchange purification) for evaluation for the first time at -20 °C.
- Re-suspend the cells from 1 L cell culture thoroughly with 40 ml of PSP buffer in a 100 ml glass beaker.
- Anion exchange purification
- Set up the FPLC machine and turn on the 280 nm UV lights for detection.
- Install a 5 ml Hitrap FF Q column on the FPLC machine, and wash it with 25 ml of ddH2O and then 25 ml of buffer A at 4.0 ml/min.
- Exchange buffer of the KaiC protein from the last step with buffer A using a NAP-25 buffer exchange column, and load the sample onto the column at 2.0 ml/min.
- Wash the column with buffer A until the absorbance at 280 nm reaches the baseline.
- Wash the column with 10% buffer B (90% buffer A) to remove non-specifically bound proteins.
- Set up a linear gradient elution program with 10-50% buffer B for 70 min, and collect the protein peak (~ 40 min) which contains KaiC.
- Keep the KaiC sample at 4 °C.
- All buffers and samples used for FPLC purification must be filtered through 0.22 µm filters.
- The linear gradient elution step is strongly recommended for the first purification trial.
- Set up the FPLC machine and turn on the 280 nm UV lights for detection.
- Storage of KaiC
- To store the purified KaiC protein, concentrate the KaiC sample from the last step (step D7) to 1-5 mg/ml with a 10 kDa centrifugal filter at 4 °C, 3,000 rpm.
- Exchange buffer with reaction buffer using a NAP-5 buffer exchange column. Concentrate the KaiC sample to 1-5 mg/ml, and store the protein at -80 °C.
- Determine the concentration of the sample with the Bradford assay.
- We suggest using the protein immediately for the following procedures. For storage add 20% glycerol, freeze in liquid nitrogen and keep at -80 °C. Please note that long-term storage (2-3 months) might result in an activity loss of KaiC.
- To store the purified KaiC protein, concentrate the KaiC sample from the last step (step D7) to 1-5 mg/ml with a 10 kDa centrifugal filter at 4 °C, 3,000 rpm.
- Preparation of KaiC samples for SDS-PAGE analysis
- Take out the KaiC protein stored at -80 °C and thaw on ice.
- Dilute KaiC to 0.5 mg/ml with reaction buffer supplemented with 80 µg/ml of kanamycin to a final volume of 30 µl.
- Transfer 4 µl of the diluted KaiC sample to clean and sterilized PCR tubes labelled as tubes 1 to 5.
- Add 2 µl of 3x loading dye to sample tube 1 and incubate at 100 °C for 5 min.
- Store sample 1 at -20 °C and transfer samples 2-5 to 4 °C.
- After 24 h, take out sample 2 and prepare as described above.
- Place the sample tubes 3-5 in a PCR machine, and set the sample temperature to 30 °C.
- Take out one tube every 4 h and prepare as described before.
- Take out the KaiC protein stored at -80 °C and thaw on ice.
- SDS-PAGE analysis of the auto-phosphatase activity of KaiC
- Take out the KaiC protein stored at -80 °C and thaw on ice.
- Dilute KaiC to 0.5 mg/ml with reaction buffer supplemented with 80 µg/ml of kanamycin to a final volume of 30 µl.
- Transfer 4 µl of the diluted KaiC sample to clean and sterilized PCR tubes labelled as tubes 1 to 5.
- Add 2 µl of 3x loading dye to sample tube 1 and incubate at 100 °C for 5 min.
- Store sample 1 at -20 °C and transfer samples 2-5 to 4 °C.
- After 24 h, take out sample 2 and prepare as described above.
- Place the sample tubes 3-5 in a PCR machine, and set the sample temperature to 30 °C.
- Take out one tube every 4 h and prepare as described before.
- The best amount of KaiC in each well is 2-3 µg in this protocol.
- Add 10 µl of the loading dye in the wells beside the samples to reduce the ‘smile effect’.
- Take out the KaiC protein stored at -80 °C and thaw on ice.
Data analysis
- Evaluation of KaiC purity
Since protein purity is important for the activity and stability of proteins, check the purity of the purified KaiC protein by SDS-PAGE. As shown in Figure 1, the KaiC prepared with this protocol was highly pure, and the molecular weight corresponded to the predicted value of 58 kDa.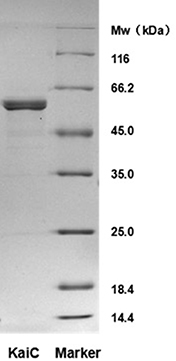
Figure 1. SDS-PAGE analysis of purified KaiC - Analysis of auto-phosphatase activity of KaiC
To determine the auto-phosphatase activity of the purified KaiC, the samples prepared at different time points (1-5) were analyzed by SDS-PAGE. Figure 2 clearly shows the de-phosphorylation of KaiC when incubated alone at 30 °C in reaction buffer. The lower band is the de-phosphorylated KaiC, and the upper band is the phosphorylated KaiC.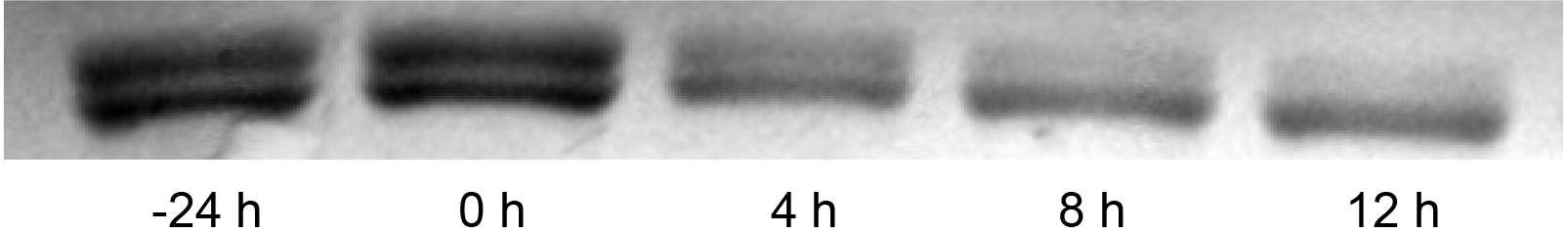
Figure 2. SDS-PAGE analysis of auto-phosphatase activity of KaiC. KaiC was incubated at 4 °C for 24 h (-24 h) before incubation at 30 °C. Samples were taken every 4 h. - Quantitation of KaiC auto-phosphatase activity
To quantitatively analyze the de-phosphorylation of KaiC, the SDS-PAGE gel was analyzed with ImageJ (Schneider et al., 2012). Open the recorded high-quality image of the SDS-PAGE gel in ImageJ. Frame the KaiC bands in the first lane, and select ‘Analyze-Gels-Select First Lane’. Move the frame to the next lane while keeping its size unchanged to cover the KaiC bands, and then use the ‘Analyze-Gels-Select Next Lane’ function. Repeat this process for all lanes. Use the function ‘Analyze-Gels-Plot Lanes’ to plot the peak areas of the KaiC bands. Then draw the baselines with the line tool to close up the independent peak areas corresponding to the KaiC bands in the gel. Use the ‘Wand Tool’ to select the closed peak areas one by one. A new window will show the calculated peak areas. Finally, calculate the percentages of the phosphorylated KaiC band in each lane, and analyze the data in Excel or other similar software (Figure 3).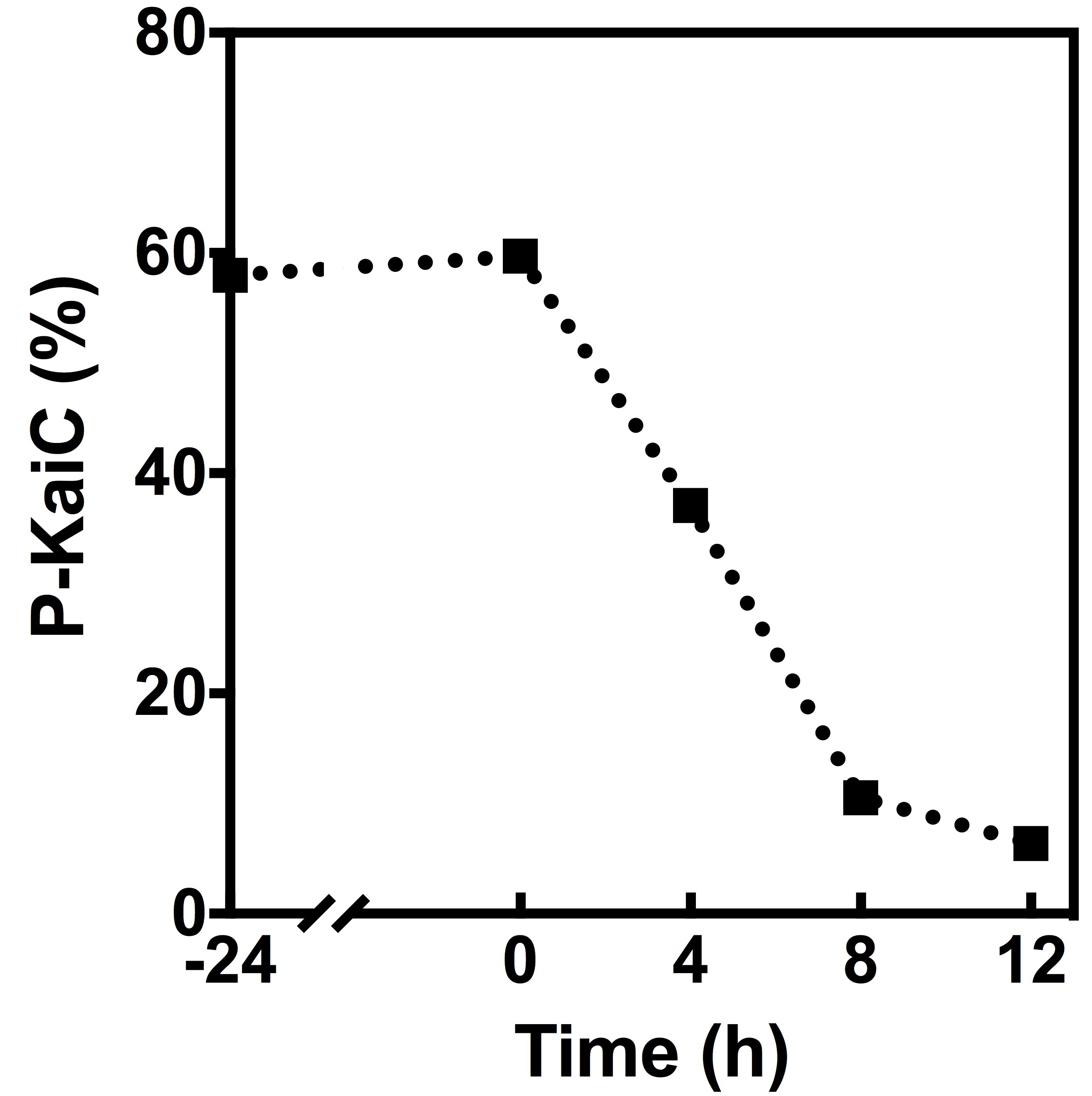
Figure 3. Quantitative analysis of the auto-phosphatase activity of KaiC. The percentage of the phosphorylated KaiC (P-KaiC) in each lane was analyzed by ImageJ.
Note: The baseline correction in ImageJ is somehow arbitrary, but it should be kept constant for all peaks.
Recipes
- PSP buffer
50 mM Tris-HCl (pH 8.0)
150 mM NaCl
1 mM DTT
0.01 % Tween-20
1 mM ATP
5 mM MgCl2 - Buffer A
50 mM Tris-HCl (pH 8.0)
1 mM DTT
0.01 % Tween-20
1 mM ATP
5 mM MgCl2 - Buffer B
50 mM Tris-HCl (pH 8.0)
1 M NaCl
1 mM DTT
0.01 % Tween-20
1 mM ATP
5 mM MgCl2 - Reaction buffer
50 mM Tris-HCl (pH 8.0)
150 mM NaCl
5 mM ATP
5 mM MgCl2
0.01 % Tween-20
0.5 mM EDTA - 1 M IPTG
Dissolve 1 g IPTG in 4,196 μl deionized water to make 1 M solution
Filter sterilize with syringe and 0.22 μm filter - 10x running buffer
144 g of glycine
30.3 g of Tris
10 g of SDS
Add ddH2O to 1 L - 8% separation gel (Table 1)
Table 1. Preparation of the 8% separation gel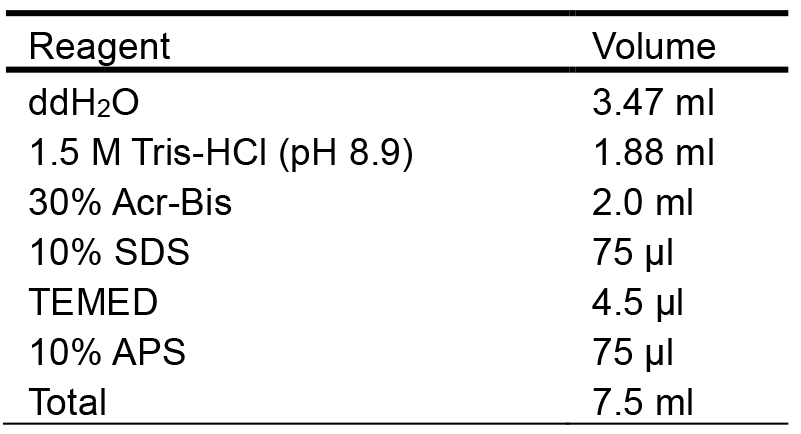
- 5% stacking gel (Table 2)
Table 2. Preparation of the 5% stacking gel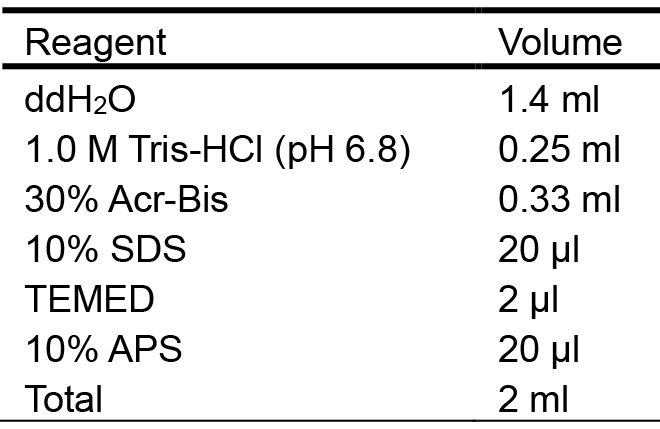
- Coomassie blue staining solution (1 L)
1 g Coomassie blue G250
650 ml ddH2O
250 ml methanol
100 ml acetic acid - De-staining buffer (1 L)
600 ml ddH2O
300 ml methanol
100 ml acetic acid
Notes:
- All buffers should be sterilized with 0.45 µm filters.
- Store buffers at 4 °C for less than a week.
Acknowledgments
This work was funded by the grants to S.L. from the National Natural Science Foundation of China (91330113, 31670768), Hubei Province of China (D20161204), and China Three Gorges University. The reference work was published in Sci Rep 6: 25129.
References
- Chen, W., Liu, S. and Liu, S. (2013). Advances in the molecular mechanism of the core circadian oscillator of cyanobacteria. Acta Biophysica Sinica 29(11): 801-810.
- Dong, P., Fan, Y., Sun, J., Lv, M., Yi, M., Tan, X. and Liu, S. (2016). A dynamic interaction process between KaiA and KaiC is critical to the cyanobacterial circadian oscillator. Sci Rep 6: 25129.
- Egli, M. and Johnson, C.H. (2013). A circadian clock nanomachine that runs without transcription or translation. Curr Opin Neurobiol 23(5): 732-740.
- Egli, M. (2015). Structural and biophysical methods to analyze clock function and mechanism. Methods Enzymol 551: 223-266.
- Johnson, C. H., Stewart, P. L. and Egli, M. (2011). The cyanobacterial circadian system: from biophysics to bioevolution. Annu Rev Biophys 40: 143-167.
- Mackey, S. R., Golden, S. S. and Ditty, J. L. (2011). The itty-bitty time machine genetics of the cyanobacterial circadian clock. Adv Genet 74: 13-53.
- Ma, P., Woelfle, M. A. and Johnson, C. H. (2013). An evolutionary fitness enhancement conferred by the circadian system in cyanobacteria. Chaos Solitons Fractals 50: 65-74.
- Nakajima, M., Imai, K., Ito, H., Nishiwaki, T., Murayama, Y., Iwasaki, H., Oyama, T. and Kondo, T. (2005). Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308(5720): 414-415.
- Nishiwaki, T. and Kondo, T. (2012). Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem 287(22): 18030-18035.
- Pattanayak, G. and Rust, M. J. (2014). The cyanobacterial clock and metabolism. Curr Opin Microbiol 18: 90-95.
- Roenneberg, T. and Merrow, M. (2016). The circadian clock and human health. Curr Biol 26(10): R432-443.
- Rust, M. J. (2012). Orderly wheels of the cyanobacterial clock. Proc Natl Acad Sci U S A 109(42): 16760-16761.
- Schneider, C. A., Rasband, W. S. and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7): 671-675.
- Shi, S. Q., Ansari, T. S., McGuinness, O. P., Wasserman, D. H. and Johnson, C. H. (2013). Circadian disruption leads to insulin resistance and obesity. Curr Biol 23(5): 372-381.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chen, Q., Yu, L., Tan, X. and Liu, S. (2017). Expression and Purification of Cyanobacterial Circadian Clock Protein KaiC and Determination of Its Auto-phosphatase Activity. Bio-protocol 7(4): e2140. DOI: 10.21769/BioProtoc.2140.
Category
Microbiology > Microbial biochemistry > Protein > Activity
Microbiology > Microbial biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link













