- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Aggregation Prevention Assay for Chaperone Activity of Proteins Using Spectroflurometry
(*contributed equally to this work, §Deceased) Published: Vol 7, Iss 2, Jan 20, 2017 DOI: 10.21769/BioProtoc.2107 Views: 12682
Reviewed by: Valentine V TrotterSoazig Le Guyon Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Large-scale Purification of Type III Toxin-antitoxin Ribonucleoprotein Complex and its Components from Escherichia coli for Biophysical Studies
Parthasarathy Manikandan [...] Mahavir Singh
Jul 5, 2023 2169 Views

Isolation of Antigen-Specific Nanobodies From Synthetic Libraries Using a Protein Selection Strategy That Combines MACS-Based Screening of YSD and FLI-TRAP
Apisitt Thaiprayoon [...] Dujduan Waraho-Zhmayev
Jan 20, 2026 443 Views
Abstract
The ability to stabilize other proteins against thermal aggregation is one of the major characteristics of chaperone proteins. Molecular chaperones bind to nonnative conformations of proteins. Folding of the substrate is triggered by a dynamic association and dissociation cycles which keep the substrate protein on track of the folding pathway (Figure 1). Usually molecular chaperones exhibit differential affinities with different conformations of the substrate. With the exception of the sHsp family of molecular chaperones, the shift from a high-affinity binding state to the low-affinity release state is triggered by ATP binding and hydrolysis (Haselback and Buchner, 2015). Aggregation prevention assay is a simple, yet definitive assay to determine the chaperone activity of heat labile proteins such as Maltodextrin glucosidase (MalZ), Citrate Synthase (CS) and NdeI. This is based on the premise that proteins with chaperone like activity should prevent protein substrates (MalZ, CS and NdeI) from thermal aggregation. Here, we describe a detailed protocol for aggregation prevention assay using two different chaperone proteins, resistin and MoxR1, identified from our lab. Resistin, a human protein (hRes) and MoxR1 a Mycobacterium tuberculosis protein were analysed for their effect on prevention of MalZ/Citrate Synthase (CS)/NdeI aggregation.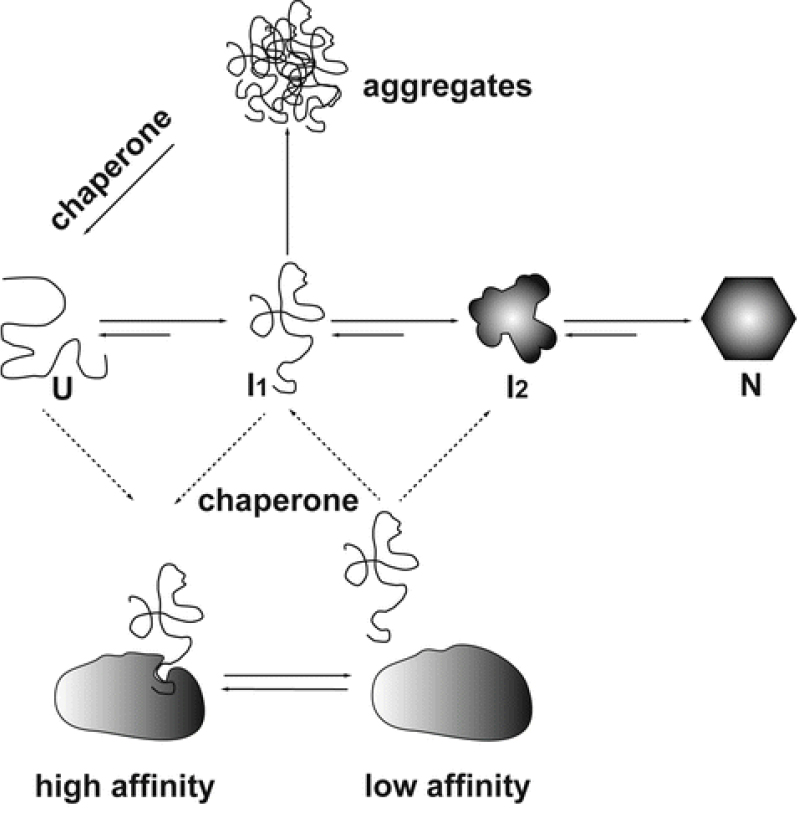
Figure 1. Mechanism of action of molecular chaperones. Citrate synthase folds via increasingly structured intermediates (I1, I2) from the unfolded state (U) to the folded state (N). Under heat shock conditions, this process is reversed.
Background
To elucidate the chaperone activity of a protein in vitro, several methods have been developed. Primarily these methods are based on examining the enzyme function and the ability of the chaperone to refold and protect enzyme activity under heat or other stress conditions. Other method to identify and study chaperones includes in silico analysis, or co-purification with other proteins. The limitations with such methods are less reproducibility or inherent high chances of false positive results. In the current method the use of light scattering to detect prevention of protein aggregation relies on thermal stabilization of protein only from denatured state, and in the presence of a chaperone. In contrast, if aggregate is formed from native or intermediate state of the protein the amount of aggregation might increase thereby decreasing the chance of false positive. Furthermore, this method uses the purified recombinant protein for the assay and therefore, can also be used to study other chaperone proteins from other bacterial sources.
Materials and Reagents
- Pipette tips
- Microcentrifuge tube
- Parafilm
- E. coli BL21 (DE3) cells
- Citrate synthase ammonium sulfate suspension from porcine heart (Sigma-Aldrich, catalog number: C3260 )
- Ammonium sulphate
- Tris (pH 8.0) (AMRESCO, catalog number: 0497 )
- Luria Bertani agar plates
- Ampicillin (Sigma- Aldrich, catalog number: A9518 )
- Isopropyl β-D-1-thiogalactopyranoside, IPTG (Sigma-Aldrich, catalog number: I6758 )
- DNAse I (Sigma-Aldrich, catalog number: AMPD1 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M2670 )
- Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: 7626 )
- Bradford reagent (Bio-Rad Laboratories, catalog number: 5000006 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A7906 )
- Recombinant protein MoxR1 and hRes (1 mg/ml) (Purified in the laboratory)
- Purified recombinant protein GroEL (0.5 mg/ml) (Purified in the laboratory)
- Lysozyme (1 mg/ml) (Sigma-Aldrich, catalog number: 4919 )
- Milli-Q water
- NdeI (20,000 U/ml) (New England BioLab, catalog number: R0111 )
- Maltodextrin glucosidase protein (1 mg/ml) (Purified in the laboratory)
- EDTA (Sigma-Aldrich, catalog number: E9884 )
- Sodium phosphate (dibasic) heptahydrate (Na2HPO4·7H2O) (AMRESCO, catalog number: 0348 )
- Sodium phosphate (monobasic) anhydrous (NaH2PO4) (AMRESCO, catalog number: 0571 )
- Sodium chloride (NaCl) (AMRESCO, catalog number: 0241 )
- Imidazole (AMRESCO, catalog number: 0527 )
- Glycerol (AMRESCO, catalog number: 0854 )
- TE buffer (see Recipes)
- 20 mM sodium phosphate buffer pH 7.4 (see Recipes)
- Binding buffer (see Recipes)
- Washing buffer (see Recipes)
- Elution buffer (see Recipes)
- Dialysis buffer (see Recipes)
Equipment
- Pipette (Gilson, model: Pipetman Neo®)
- Centrifuge (Hermle Labor Technik, model: Z 326 K )
- PerkinElmer spectrofluorometer (PerkinElmer, model: LS55 ) with controlled temperature peltier block
- Incubator (Eppendorf, BrunswickTM, model: 44/44R )
- Quartz cuvette (PerkinElmer, Suprasil®, catalog number: B0631071 )
- pH meter (Thermo Fisher Scientific, Thermo Scientific, model: CyberScan Ph 510 )
- Fluorescence spectrophotometer (Agilent Technologies, model: Cary Eclipse )
Software
- Microsoft Excel
- Graphpad Prism 5
Procedure
- Preparation of citrate synthase and maltodextrin glucosidase
- Commercially available citrate synthase is supplied in ammonium sulphate suspension to keep the enzyme in an inactive state. The suspension is then pipetted into a fresh microcentrifuge tube and centrifuge at 15,000 x g, for about 10 min at 4 °C. The enzyme CS is present in a solid state and therefore will be collected in the pellet fraction.
- Discard the supernatant and dissolve the pellet containing enzyme CS in sterile water at a concentration of 1 mg/ml.
- The CS should be further purified by size exclusion chromatography and dialysed against 50 mM Tris (pH 8.0). For refolding assays, CS stock solutions at concentration of 150 μM is prepared in TE buffer by concentrating CS using Amicon Ultra microconcentrator with 30 kDa cutoff before use.
- The expression plasmids pCS19MalZ containing (His)6 tagged coding region of malz gene are transformed into E. coli BL21 (DE3) competent cells and plating is performed on Luria Bertani agar plates containing 100 µg/ml ampicillin for selecting pCS19MalZ.
- A single colony from the transformed bacterial cells grown on Luria Bertani agar plate containing 100 µg/ml ampicillin is picked and grown in the Luria Bertani broth containing 100 µg/ml ampicillin.
- The tube is incubated for overnight at 37 °C with constant shaking at a rate of 150 rpm.
- About 2% of the overnight culture is inoculated in fresh Luria Bertani broth containing ampicillin 100 µg/ml and then incubate it for 2 h at 37 °C with constant shaking at a rate of 150 rpm.
- The expression of MalZ protein is then induced with 100 µM IPTG and left for 12 h.
- The IPTG induced culture is pelleted through centrifugation at 11,510 x g for 50 min. The supernatant is discarded, wash the cell pellet with binding buffer and resuspend the cells in binding buffer, along with DNAse I (1 µl/ml of cell lysate), 0.5 mM MgCl2 and 1 mM PMSF (a serine protease inhibitor).
- The cells are disrupted through sonication on ice, and the supernatant is collected after centrifugation. (Sonication was performed for 5 min at 1 sec on and 2 sec off cycle and at 30% amplitude)
- The protein is further purified by Ni-NTA affinity chromatography. The column is washed with 5 column volumes of washing buffer and the protein is eluted with 5 column volumes of elution buffer (Goyal et al., 2014).
Note: Nickel nitrilotriacetic [Ni-NTA] chromatography is used for purification of 6x His-tagged recombinant proteins overproduced in bacteria. The Ni-NTA agarose resins possess high affinity and specificity for 6x His-tagged recombinant fusion proteins. The proteins bound to the resin are eluted by competition with imidazole. - The concentration of the protein is determined using Bradford reagent. The standard curve is plotted using different concentration of bovine serum albumin by determining the Optical Density using spectrophotometer and plotting the 595 nm values (y-axis) versus their concentration in µg/ml (x-axis).The concentration of dialyzed protein is then determined using the standard curve.
- Commercially available citrate synthase is supplied in ammonium sulphate suspension to keep the enzyme in an inactive state. The suspension is then pipetted into a fresh microcentrifuge tube and centrifuge at 15,000 x g, for about 10 min at 4 °C. The enzyme CS is present in a solid state and therefore will be collected in the pellet fraction.
- Thermally induced aggregation assay
- To elucidate the chaperonic function of hRes protein on the thermal aggregation of CS, recombinant GroEL protein is used as a positive control and lysozyme as a negative control.
Note: Chaperone to substrate ratio is critical and initially different ratios should be used to determine the optimal ratio for the effect of chaperone activity. - We maintained 1:1 ratio between hRes (chaperone) vs. CS (Suragani et al., 2013). The proteins are used at 0.15 µM concentration.
- Similarly, to analyze the chaperone function of MoxR1 protein on thermal aggregation of MalZ, GroEL is used as a positive control and lysozyme as a negative control. GroEL and lysozyme are used at 2 µM concentration. In this case, we maintained the initial 12:1 ratio between MoxR1 (chaperone) and MalZ (Bhuwan et al., 2016) (see Note 1).
- Stirred 0.75 ml quartz cuvettes are to be used in a fluorescence spectrophotometer. The stirring is achieved by removing the cuvettes out of the spectrophotometer and sealing it by Parafilm and mixing the solution by see-saw movement during the time interval of each data point. The emission and excitation slits should be set to 2-5 nm.
- Pre-equilibrate 50 mM Tris (pH 8.0) with and without chaperones at 45-47 °C for 50 min. The chaperone dialysis buffer without chaperones is used as a control which is included in the assay to rule out the possibility of stabilizing effects of buffer.
Note: After each sample analysis the quartz cuvettes should be thoroughly washed with a strong jerk using MilliQ water to remove the traces of previous protein samples and any film formed on the side of the cuvettes and after washing, the cuvette should be dried before using it at the next step. - The initial light scattering signals are monitored at a constant wavelength of 320 nm for CS and NdeI. Here, the data points should be taken at every 5 min and the kinetics are constantly measured for 45 min at 45 °C. The increase in light scattering is observed to visualize the thermal aggregation of CS.
- The light scattering signal for MalZ is to be recorded at 500 nm wavelength.
Note: The range of absorbance for dynamic light scattering to detect aggregation is 300 nm to 600 nm and can also be optimized for different chaperone analysis. - Take the data at 5 min interval and the kinetics is monitored for 15 min at 47 °C (see Note 2).
- The signal prior to CS or NdeI and MalZ addition is set to 0 (= no aggregation).
- The readings are exported in a Microsoft Excel format and graph.
- Plot the data for Optical Density on x-axis vs. Time on y-axis using Origin or Graphpad Prism 5 software to obtain a graph.
- The resulting sigmoidal graphs will allow identifying the chaperonic function of a protein by preventing thermal aggregation of CS or NdeI and MalZ.
- To elucidate the chaperonic function of hRes protein on the thermal aggregation of CS, recombinant GroEL protein is used as a positive control and lysozyme as a negative control.
Data analysis
The experimental data showing recombinant hRes and MoxR1 proteins mediated prevention of thermal aggregation of CS or NdeI and MalZ proteins are depicted in Figures 2 and 3.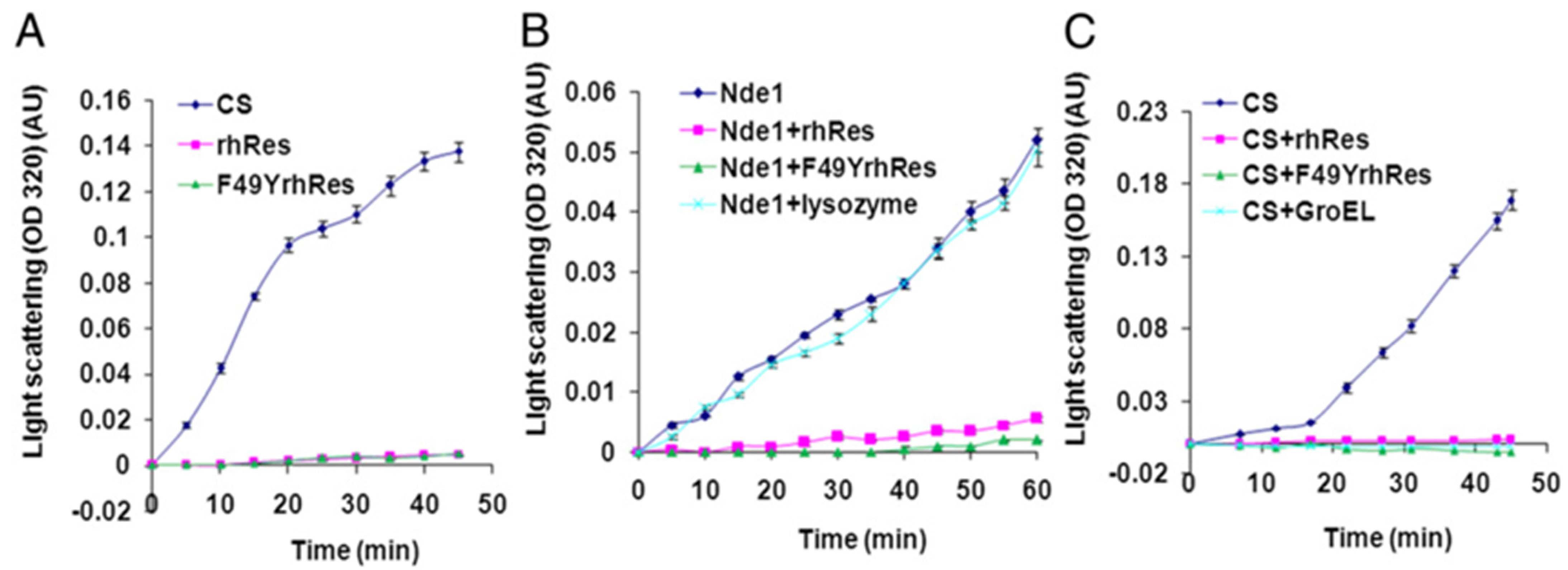
Figure 2. rhRes, proteins prevent thermal aggregation of citrate synthase (CS) and NdeI proteins. Light scattering assay for aggregation prevention for (A) rhRes, F49YrhRes, and CS at 45 °C for 45 min. B. Resistin prevents thermal aggregation of NdeI and C. Citrate synthase. Each experiment should be performed in triplicates (Suragani et al., 2013). GroEL served as positive control while lysozyme was used as a negative control. F49 amino acid of hRes predicted to be an important residue to exhibit chaperonic activity was changed to tyrosine by site directed mutagenesis.
Note: Proteins such as CS consist of polypeptide chains that are affected by multiple parameters such as temperature, pH and chemical environment. Further, preparation method, storage conditions or buffer varies from batch to batch; these can influence the scattering pattern of the proteins in a sample. 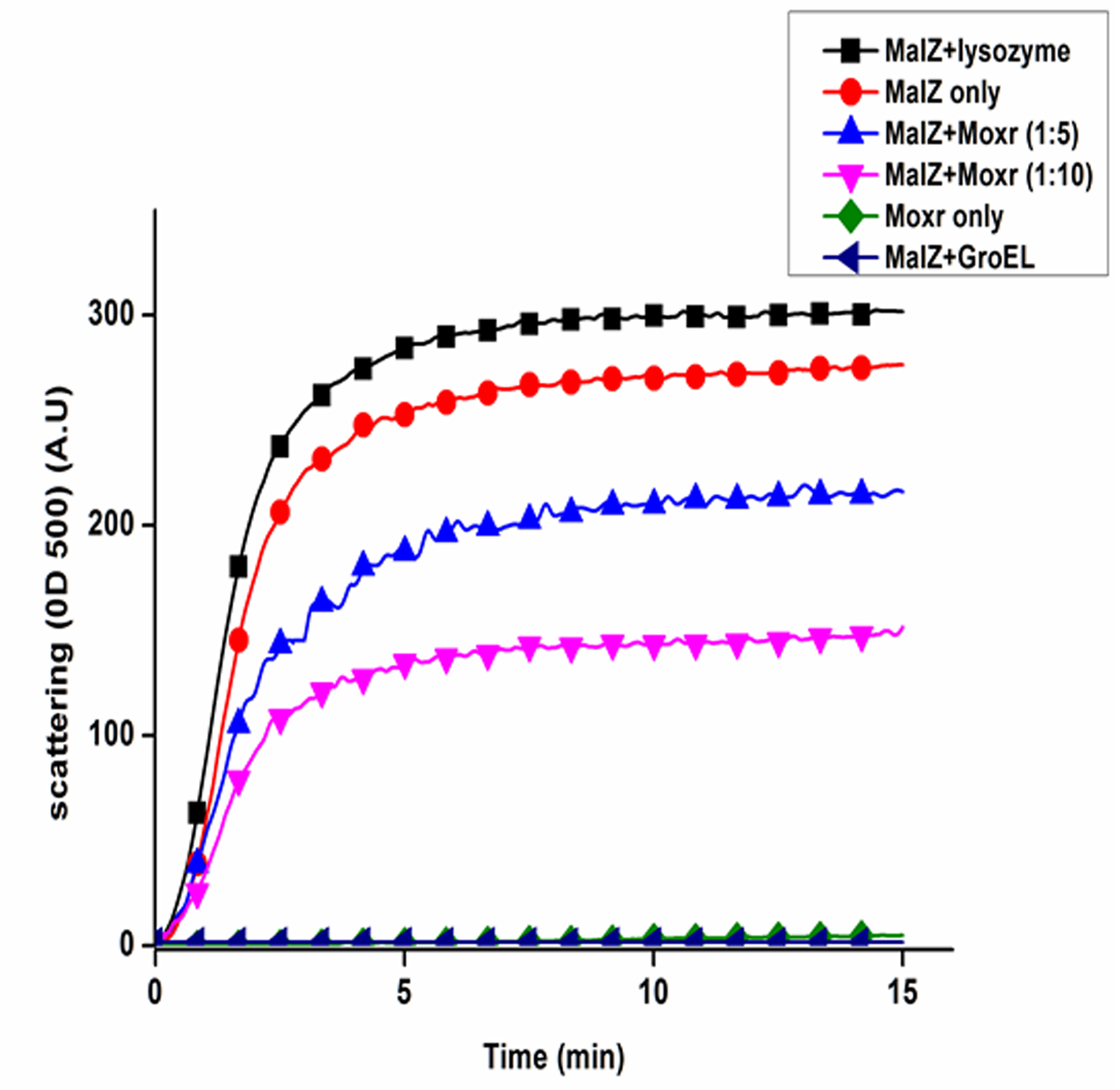
Figure 3. MoxR1 and GroEL proteins prevent thermal aggregation of MalZ protein. MoxR1 and GroEL were used as positive control while lysozyme was used as a negative control. Each experiment should be performed in triplicate (Bhuwan et al., 2016).
Notes
- To check the activity of chaperones on the inactivation of CS or other heat liable proteins such as MalZ, a different range of stoichiometric ratios up to 32:1 (chaperone to CS) can be used (Haselback and Buchner, 2015)
- Since the slope at the beginning of the reaction is used for comparison, it is important that both pipetting as well as sample processing is always performed simultaneously.
Recipes
Note: Buffer composition for MalZ protein purification.
- TE buffer
50 mM Tris pH 8.0
2 mM EDTA pH 8.0
Autoclaved at 121 °C for 20 min - 200 mM sodium phosphate buffer pH 7.4 (50 ml)
Mix 40.5 ml of 0.2 M Na2HPO4·2H2O and 9.5 ml of 0.2 M NaH2PO4·H2O
0.2 M Na2HPO4·2H2O (35.6 g/L)
0.2 M NaH2PO4·H2O (27.6 g/L) - Binding buffer
20 mM sodium phosphate, containing 500 mM NaCl, at pH 7.4 - Washing buffer
20 mM sodium phosphate, containing 500 mM NaCl and 10 mM imidazole, at pH 7.4 - Elution buffer
20 mM sodium phosphate, containing 500 mM NaCl and 500 mM imidazole, at pH 7.4 - Dialysis buffer
20 mM sodium phosphate, containing 500 mM NaCl and 10% glycerol, at pH 7.4
Acknowledgments
This protocol was adapted from our earlier publications (Suragani et al., 2013 and Bhuwan et al., 2016). The work was supported by a Centre of Excellence Phase 2 Grant (BT/R12817/COE/34/23/2015) from the Department of Biotechnology, Ministry of Science & Technology, Government of India to S.E.H. and N.Z.E.
Competing interests
The authors declare no conflict of interest or competing interest.
References
- Bhuwan, M., Arora, N., Sharma, A., Khubaib, M., Pandey, S., Chaudhuri, T. K., Hasnain, S. E. and Ehtesham, N. Z. (2016). Interaction of Mycobacterium tuberculosis virulence factor RipA with chaperone MoxR1 is required for transport through the TAT secretion system. mBio 7(2): e02259.
- Goyal, M., Chaudhuri, T. K. and Kuwajima, K. (2014). Irreversible denaturation of maltodextrin glucosidase studied by differential scanning calorimetry, circular dichroism, and turbidity measurements. PLoS One 9(12): e115877.
- Haslbeck, M. and Buchner, J. (2015). Assays to characterize molecular chaperone function in vitro. Methods Mol Biol 1292: 39-51.
- Suragani, M., Aadinarayana, V. D., Pinjari, A. B., Tanneeru, K., Guruprasad, L., Banerjee, S., Pandey, S., Chaudhuri, T. K. and Ehtesham, N. Z. (2013). Human resistin, a proinflammatory cytokine, shows chaperone-like activity. Proc Natl Acad Sci U S A 110(51): 20467-20472.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bhuwan, M., Suragani, M., Ehtesham, N. Z. and Hasnain, S. E. (2017). Aggregation Prevention Assay for Chaperone Activity of Proteins Using Spectroflurometry. Bio-protocol 7(2): e2107. DOI: 10.21769/BioProtoc.2107.
Category
Microbiology > Microbial biochemistry > Protein > Structure
Microbiology > Microbial biochemistry > Protein > Interaction
Biochemistry > Protein > Interaction > Protein-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












