- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation of Tumour-stroma Minispheroids for Drug Efficacy Testing
Published: Vol 7, Iss 1, Jan 5, 2017 DOI: 10.21769/BioProtoc.2091 Views: 9998
Reviewed by: HongLok LungKevin Patrick O’RourkeVikash Verma

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparation of Recombinant Galectin-3 for Cancer Studies
Kari Tyler [...] Erwin G. Van Meir
Jan 5, 2016 9347 Views

Optimizing Confocal Imaging Protocols for Muscle Fiber Typing in the Mouse Masseter Muscle
Catalina Matias [...] Jeffrey J. Brault
Apr 5, 2025 2900 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 481 Views
Abstract
The three-dimensional organisation of cells in a tissue and their interaction with adjacent cells and extracellular matrix is a key determinant of cellular responses, including how tumour cells respond to stress conditions or therapeutic drugs (Elliott and Yuan, 2011). In vivo, tumour cells are embedded in a stroma formed primarily by fibroblasts that produce an extracellular matrix and enwoven with blood vessels. The 3D mixed cell type spheroid model described here incorporates these key features of the tissue microenvironment that in vivo tumours exist in; namely the three-dimensional organisation, the most abundant stromal cell types (fibroblasts and endothelial cells), and extracellular matrix. This method combined with confocal microscopy can be a powerful tool to carry out drug sensitivity, angiogenesis and cell migration/invasion assays of different tumour types.
Keywords: Mixed cell type 3-dimensional (3D) cultureBackground
The traditional monolayer cell culture (2-dimensional) enforces an artificial environment, which is vastly different from the tissues cells exists in vivo. One of the most critical differences is that in monolayer cultures the cells are polarised, i.e., the surface of the cells facing the culture-plastic and the upper cell surface exposed to the culture medium receive completely different, often opposing signals (Fitzgerald et al., 2015). To address the problem of cell polarization, tumour spheroid cultures are increasingly used in cancer research. Tumour spheroids can replicate the 3-dimensional cell-cell interactions present in a tissue and to some extent paracrine signaling via cytokines and chemokines by reducing their diffusion and dilution by the growth medium that typically occurs in monolayer cultures (Lawlor et al., 2002; Barrera-Rodríguez and Fuentes, 2015). The current tumour-stroma minispheroid protocol is one such method. Compared to the other tumour-spheroid protocols, this method also incorporates additional, key features of the tissue environment, namely stromal cells and extracellular matrix in the spheroid and thus provides a model that replicates the in vivo tumour microenvironment more faithfully.
Materials and Reagents
- 96 U-shaped well plate for suspension cells (Greiner Bio One, catalog number: 650161 )
- FiltopurTM syringe filters (SARSTEDT, catalog number: 83.1826.001 )
- 50 ml syringes (TERUMO, catalog number: SS+50ES )
- 12 well dishes with 10 mm diameter glass bottom (MATTEK, catalog number: P12G-0-10-F )
- 1.5 ml sterile Eppendorf tubes (SARSTEDT, catalog number: 72.690.001 )
- 50 ml sterile centrifuge tubes (Corning, catalog number: 430829 )
- 35 mm glass bottom dish, 14 mm diameter (MATTEK, catalog number: P35G-0.170-14-C )
- T75 flasks for adherent cells (SARSTEDT, catalog number: 83.3911 )
- Serological pipettes (5 ml, 10 ml) (CORNING, catalog numbers: 4051 and 4101 , respectively)
- Pipette Tips (10 μl, 200 μl, 1,000 μl) (SARSTEDT, catalog numbers: 70.1130.100 , 70.760.002 and 70.762.100 , respectively)
- Cell lines: MDA-MB-231 breast cancer epithelial cells (ATCC, HTB-26TM, catalog number: MDA-MB-231); human umbilical vein endothelial cells (HUVEC) (ATCC, CRL-1730TM, catalog number: HUV-EC-C ); normal human dermal fibroblasts (NHDF) (Lonza, catalog number: CC-2509 )
- Recombinant human tumour necrosis factor-related apoptosis-inducing ligand (rhTRAIL) (purified in-house), receptor-selective TRAIL mutant, TRAIL-45 (O’Leary et al., 2016; van der Sloot et al., 2006)
- Dulbecco’s modified Eagle medium (DMEM)-low glucose concentration (Sigma-Aldrich, catalog number: D6046 )
- Fetal bovine serum (Sigma-Aldrich, catalog number: F7524 )
- L-glutamine solution, 200 Mm stock (Sigma-Aldrich, catalog number: G7513 )
- 1x trypsin-EDTA buffer in HBSS
- CellTrackerTM CM-Dil Dye (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: C7001 ) or CMTPX red cell tracker dye (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: C34552 )
- Rat tail collagen type I (Corning, catalog number: 354236 )
- Hoechst33342 – 10 mg/ml solution in water (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: H3570 )
- SYTOX Green nucleic acid dye (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: S7020 )
- Endothelial cell growth medium-2 (EGM-2) prepared by adding EGMTM-2 SingleQuotsTM Kit (Lonza, catalog number: CC-4176 ) to EBM-2 basal Medium (Lonza, catalog number: CC-3156 )
- Hanks’ balanced salt solution (HBSS) (Thermo Fisher Scientific, GibcoTM catalog number: 24020117 )
- 1 N NaOH solution
- Methylcellulose solution (see Recipes)
Equipment
- HeraeusTM MegafugeTM centrifuge (15 ml, 50 ml tube) (Thermo Fisher Scientific, Thermo ScientificTM, model: 16 Centrifuge Series )
- Mammalian cell culture incubator (37 °C, 5% CO2) (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM Steri-CycleTM )
- Hemocytometer
- Pipettes (10 μl, 200 μl, 1,000 μl)
- Pipette aid
- Confocal microscopy system (AndorTM, Revolution Spinning Disk Confocal systemTM)
- High-resolution EMCCD camera (Andor iXon EM+)
- Olympus IX81 motorised inverted microscope, fitted with a variable temperature/CO2 humidified incubation chamber for live cell experiments
- Yokagawa CSU22 spinning disk confocal unit
- Magnetic stirrer
- Orbital shaker
Software
- VolocityTM software (PerkinElmer)
Procedure
- Production of multicellular, mixed cell type spheroids
- Culture MDA-MB-231 cells, HUVEC cells in EGM-2 medium and NHDF in low-glucose DMEM supplemented with 10% fetal calf serum and 2 mM L-glutamine in T-75 flasks to reach near confluency.
- Harvest MDA-MB-231 cells by trypsinization. Collect 1 x 105 cells by centrifuging at 300 x g for 5 min and resuspend them in 0.1 ml fresh growth medium (obtaining a cell concentration of 1 x 106 cells/ml).
- Label MDA-MB-231 cells with the cell tracker dye CM-Dil or CMTPX by incubating the 0.1 ml of cell suspension with 2 μM of either of the two dyes for 30 min at 37 °C in the dark, shaking every 5 to 10 min.
Note: Depending on the needs of the specific assay, you may leave the tumour cells unstained and label the non-malignant cell components using the same procedure. - Harvest HUVEC and NHDF cells by trypsinisation. Collect 1 x 105 HUVEC cells and 0.5 x 105 NHDF cells by centrifuging them at 300 x g for 5 min and resuspend the cell pellets in 0.2 ml fresh growth medium (obtaining a cell concentration of 0.5 x 106 and 0.25 x 106 cells/ml, respectively).
- Add 150 μl of HUVEC cells (7.5 x 104 cells), 150 μl of NHDF (3.75 x 104 cells) and 75 μl of MDA-MB-231 cells (7.5 x 104 cells) into 15 ml of EGM-2 medium containing 20% methylcellulose solution (see Recipes).
- Add 150 μl of the above solution to each well of a 96 U-shaped well suspension plate and incubate for 24 h at 37 °C to allow for spheroid formation.
Note: Figures 1A and 1B show the transmission light microscopic image of the single cell suspension in the U-shape well and the forming sphere (with still primarily round-shaped cells) 24 h after seeding the cells.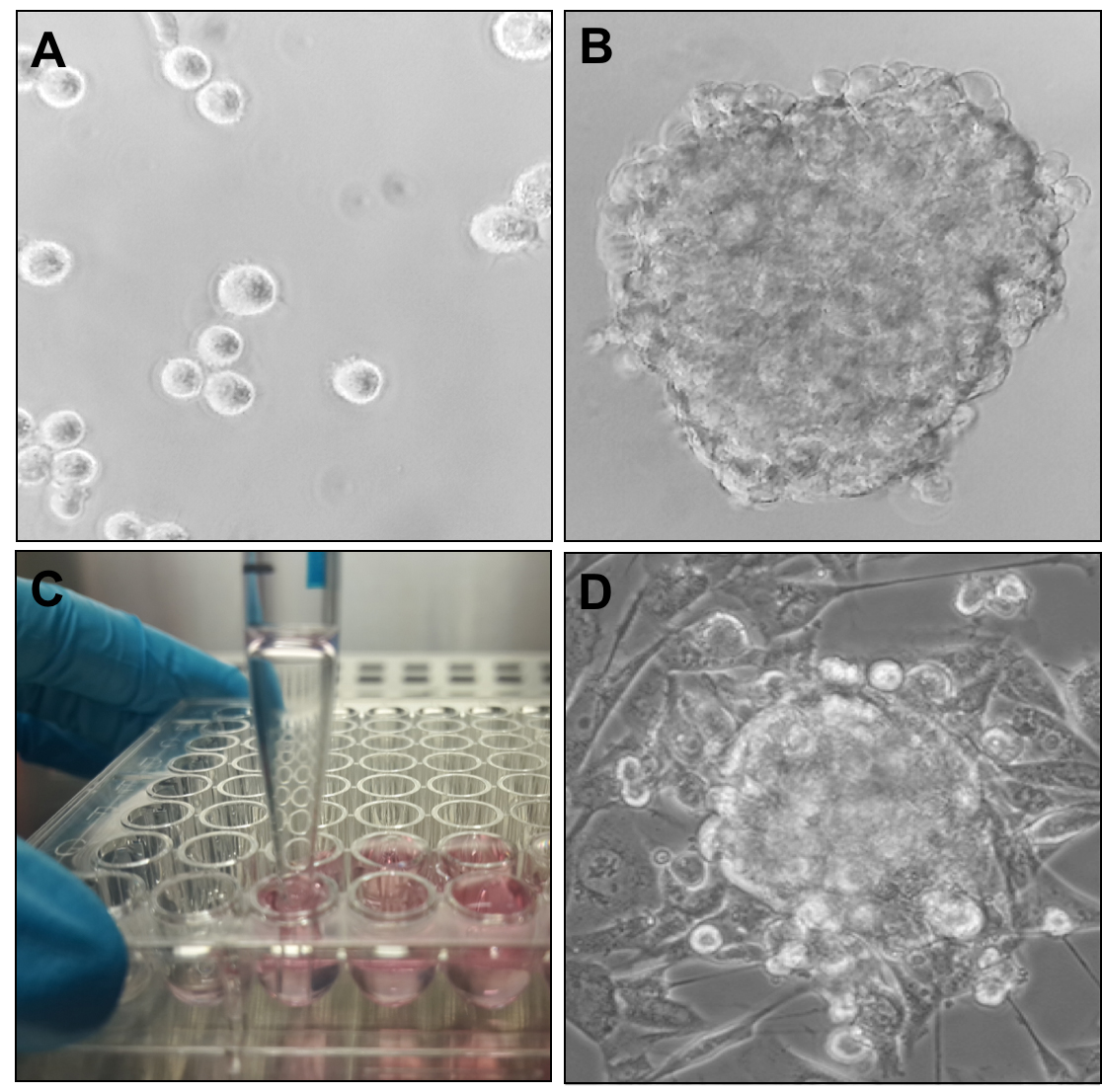
Figure 1. Formation and morphology of mixed cell type tumour minispheres. A, B and D. Transmission light microscopic images of (A) the cell constituents of the tumour sphere immediately after seeding in U-shaped wells, (B) assembled tumour sphere after 24 h of incubation in U-shaped well, and (D) fully formed tumour sphere embedded in collagen matrix at 72 h after seeding. C. Collection of formed spheres from U-shaped wells using a 5 ml serological pipette.
Note: The pipette is gently placed to the bottom of the well and the content collected. The pipette is then moved to the next well without releasing the collected medium. The medium with the tumour spheres collected in the pipette from several wells is then transferred into a 15 ml centrifuge tube (not shown).
- Culture MDA-MB-231 cells, HUVEC cells in EGM-2 medium and NHDF in low-glucose DMEM supplemented with 10% fetal calf serum and 2 mM L-glutamine in T-75 flasks to reach near confluency.
- Drug efficacy testing on multicellular tumour spheroids
- Prepare a 1.5 mg/ml collagen type-I stock solution by diluting rat tail collagen type-I into an appropriate volume of EGM-2 medium. Neutralize the pH to 7.0 by drop-wise addition of 1 N NaOH. Filter-sterilise the final solution using a syringe and syringe filter. Prepare this solution fresh each time.
- Add 100 μl of collagen stock solution to the bottom of wells of a glass-bottom 12 well dish which has been previously warmed to 37 °C in the incubator. Incubate dish at 37 °C for 30 min to allow the collagen gel to set.
- At the end of the 24 h incubation, gently harvest the formed spheroids from the 96 U-shaped well plate into a 15 ml centrifuge tube using a 5 ml or a 10 ml serological pipette and collect the spheroids by centrifuging the solution at 300 x g for 5 min.
Note: An image of the harvesting step is shown in Figure 1C. Typically, one sphere forms per well, though it may vary for different cell types. - Remove the medium, taking care not to disturb the spheroid pellet. Add 1,400 μl freshly prepared 1.5 mg/ml collagen stock solution and carefully resuspend the spheroid pellet by tapping the bottom of the tube.
- Add 100 μl of spheroid suspension on top of each collagen gel in the 12 well plate to generate between 5-8 spheroids per well. Place the plate in incubator at 37 °C for 1 h to allow this second collagen gel layer to set.
Note: It may take a bit longer for the gel to set. Proceed with the protocol only after the gel has set, but not later than 2.5 h. - Add 1.5 ml of EGM-2 medium to each well and incubate the spheroids for 24-48 h.
Note: A transmission light microscopic image of the fully formed spheroid embedded in collagen is shown in Figure 1D. - Treat cells with a drug/drugs of choice at appropriate concentration.
- 2 h before analysis, add 1 μg/ml Hoechst33342 nuclear dye and SYTOX Green viability dye at a final concentration of 1 μM to the media and complete the incubation time at 37 °C (for 2 h).
- Analyse induction of cell death by monitoring the number of SYTOX Green positive cells both in the CM-DiI/CMTPX-labelled tumour cell population and the CM-DiI/CMTPX-negative non-malignant cellular components using confocal microscopy (e.g., Taking images in 0.5 μm Z-stack planes without fixing the spheroids beforehand).
- Prepare a 1.5 mg/ml collagen type-I stock solution by diluting rat tail collagen type-I into an appropriate volume of EGM-2 medium. Neutralize the pH to 7.0 by drop-wise addition of 1 N NaOH. Filter-sterilise the final solution using a syringe and syringe filter. Prepare this solution fresh each time.
Data analysis
This protocol describes the generation of tumour spheres that can be used to assess efficacy of cytotoxic, anti-angiogenic, cytostatic as well as other drugs. Data analysis and statistics depends on the downstream applications, such as methods of detection of cell death. The above example we show is for the assessment of cytotoxicity based on microscopic detection of dying cells. The steps of the analysis are summarized below:
- 40 μl of the collagen-I solution was added in 35 mm Petri dishes with 14 mm glass slide bottom for microscopy (MatTek Corporation), which were previously warmed to 37 °C.
- The dishes were then incubated at 37 °C for 45 min to allow for the collagen gel to set.
- 4-6 spheroids harvested from the U-shape 96-well plates in a volume of 50 µl collagen-I were added in each dish on top of the collagen gel.
- The dishes were placed in the incubator at 37 °C for an additional 1-2 h to allow the second collagen gel embedding the tumour spheres to set.
- 2 ml of EGM-2 medium was then added to each well and the spheres were incubated for 48 h.
- The spheres were treated with a receptor-selective mutant of the death ligand cytokine, TRAIL, named TRAIL45a at a concentration of 250 ng/ml for 24 h.
- 2 h before analysis 1 µg/ml of Hoechst33342 and 1 µM of SYTOX Green was added to the culture media to label nuclei and dying cells, respectively.
- Induction of cell death was determined by counting the SYTOX Green positive dying/dead cells with confocal microscopy without any processing of the spheres. The steps of the detection are detailed below.
Note: Images of the spheres were taken in situ, in the glass-bottom dishes without digestion of the collagen matrix, enzymatic (or other) dissociation of the spheres or fixation with formaldehyde or other fixatives. - Images were taken from unfixed spheres using an AndorTM Revolution Spinning Disk Confocal systemTM with the following components: Yokagawa CSU22 spinning disk confocal system, four solid state laser lines: 405 nm, 488 nm, 561 nm and 640 nm; high-resolution EMCCD camera (Andor iXon EM+), Olympus IX81 motorised inverted microscope fitted with a variable temperature/CO2 humidified incubation chamber for live cell experiments. The following filters were used for the detection of the fluorescent signals: 360-390 nm/420-460 nm excitation/emission filter for Hoecsht33342, 470-495 nm/510-550 nm excitation/emission filter for SYTOX Green and 540-550 nm/575-625 nm excitation/emission filter for CMTPX.
- Z-stack images of the middle section of the spheroids were taken as 0.5 μm slices at an overall magnification of 600x (with a 60x immersion oil objective), which were then used to generate the 3D composite images shown in Figure 2 with the VolocityTM software (Perkin-Elmer).
Representative data
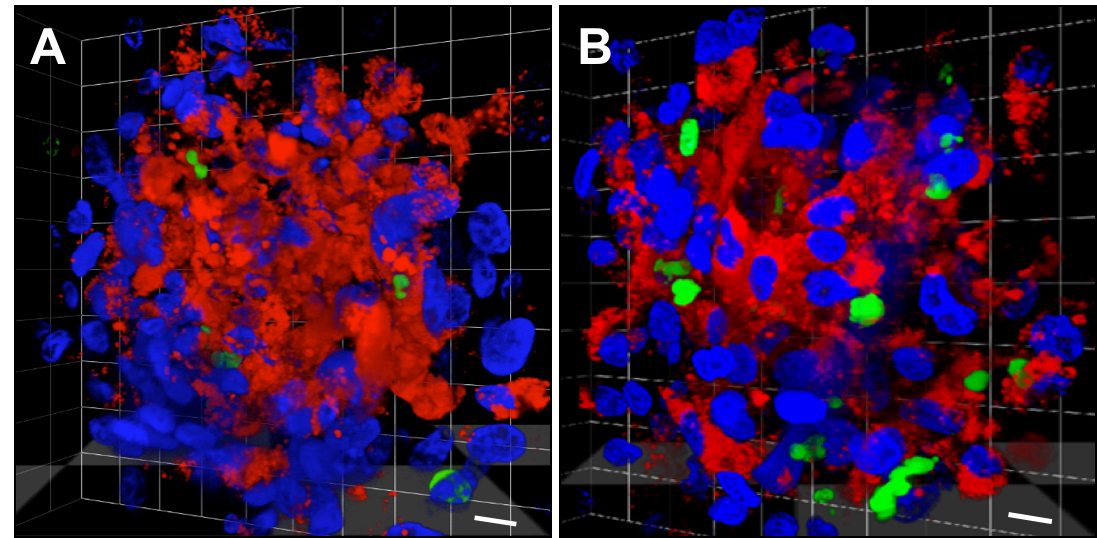
Figure 2. Detection of drug efficacy in mixed cell type tumour minispheres. 3D reconstituted confocal microscopic image of the middle 10 μm section of an (A) untreated tumour minisphere and (B) a sphere treated with an engineered, receptor-specific mutant of recombinant human TRAIL (DR4 and DR5-bispecific; TRAIL-45a). Blue: all nuclei (Hoechst33342), red: non-malignant cells: HUVEC and NHDF (CMTPX), green: dead cells (SYTOX Green). The scale bar represents 10 μM. Tumour minispheres were grown for 48 h before exposure to 250 ng/ml of TRAIL-45a for 24 h. The treated samples were stained with Hoechst33342 and SYTOX Green for the final 2 h of the treatment. Images were taken from unfixed samples using an AndorTM Revolution Spinning Disk Confocal systemTM with images taken as 0.5 μm Z-stacks at 600x magnification.
Notes
Tumour cell types other than MDA-MB-231 cells may be used, but their ability/tendency to form mixed-cell type spheroids can vary significantly and thus requires testing. Other cell types used commonly in minispheroid generation include MCF-7 and BT474 human breast cancer cells (Monazzam et al., 2006). The generation of these single-cell type spheroids uses a similar method substituting the collagen solution for agarose. Other mixed cell type spheroids involve the use of colonic adenocarcinoma cell lines, namely COLO320HSR and SNU-C1 (Park et al., 2016) mixed with fibroblasts. This protocol in particular used rotating conditions via orbital shaker to induce the generation of spheroids.
The spheroids described in this protocol can also be used to monitor angiogenesis (Correa de Sampaio et al., 2012) or cell migration/invasion.
Recipes
- Methylcellulose solution
- Autoclave 6 g of methylcellulose in a 500 ml flask with a magnetic stirrer
- Add 250 ml of EGM-2 medium previously heated to 60 °C to the autoclaved methylcellulose
- Stir to facilitate dissolving the methylcellulose for 20 min
- Add an additional 250 ml of EGM-2 medium warmed to room temperature
- Stir at 4 °C for 2 h to fully dissolve methylcellulose
- Centrifuge the final solution at 5,000 x g for 2 h at room temperature to remove undissolved methylcellulose
- Transfer the supernatant into a clean bottle
- Autoclave 6 g of methylcellulose in a 500 ml flask with a magnetic stirrer
Acknowledgments
Research in the ES laboratory is supported by Science Foundation Ireland and the Irish Cancer Society (BCNI, 14/ICS/B3042). The authors thank Prof. Gillian Murphy (Cambridge University) for sharing their minitumour protocol that this current protocol is based on.
References
- Barrera-Rodríguez, R. and Fuentes, J. M. (2015). Multidrug resistance characterization in multicellular tumour spheroids from two human lung cancer cell lines. Cancer Cell Int 15: 47.
- Correa de Sampaio, P., Auslaender, D., Krubasik, D., Failla, A. V., Skepper, J. N., Murphy, G. and English, W. R. (2012). A heterogeneous in vitro three dimensional model of tumour-stroma interactions regulating sprouting angiogenesis. PLoS One 7(2): e30753.
- Elliott, N. T. and Yuan, F. (2011). A review of three-dimensional in vitro tissue models for drug discovery and transport studies. J Pharm Sci 100(1): 59-74.
- Fitzgerald, K. A., Malhotra, M., Curtin, C. M., O'Brien, F. J. and O’Driscoll, C. M. (2015). Life in 3D is never flat: 3D models to optimise drug delivery. J Controll Release 215: 39-54.
- Lawlor, E. R., Scheel, C., Irving, J. and Sorensen, P. H. (2002). Anchorage-independent multi-cellular spheroids as an in vitro model of growth signaling in Ewing tumors. Oncogene 21(2): 307-318.
- O’Leary, L., van der Sloot, A. M., Reis, C. R., Deegan, S., Ryan, A. E., Dhami, S. P., Murillo, L. S., Cool, R. H., Correa de Sampaio, P., Thompson, K., Murphy, G., Quax, W. J., Serrano, L., Samali, A. and Szegezdi, E. (2016). Decoy receptors block TRAIL sensitivity at a supracellular level: the role of stromal cells in controlling tumour TRAIL sensitivity. Oncogene 35(10): 1261-1270.
- van der Sloot, A. M., Tur, V., Szegezdi, E., Mullally, M. M., Cool, R. H., Samali, A., Serrano, L. and Quax, W. J. (2006). Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc Natl Acad Sci U S A 103(23): 8634-8639.
- Monazzam, A., Razifar, P., Simonsson, M., Qvarnstrom, F., Josephsson, R., Blomqvist, C., Langstrom, B. and Bergstrom, M. (2006). Multicellular tumour spheroid as a model for evaluation of [18F]FDG as biomarker for breast cancer treatment monitoring. Cancer Cell Int 6: 6.
- Park, J. I., Lee, J., Kwon, J. L., Park, H. B., Lee, S. Y., Kim, J. Y., Sung, J., Kim, J. M., Song, K. S. and Kim, K. H. (2016). Scaffold-free coculture spheroids of human colonic adenocarcinoma cells and normal colonic fibroblasts promote tumorigenicity in nude mice. Transl Oncol 9(1): 79-88.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Watters, M. and Szegezdi, E. (2017). Generation of Tumour-stroma Minispheroids for Drug Efficacy Testing. Bio-protocol 7(1): e2091. DOI: 10.21769/BioProtoc.2091.
Category
Cancer Biology > General technique > Tumor microenvironment > Extracellular matrix
Cell Biology > Cell imaging > Confocal microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










