- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Generation of Mitochondrial-nuclear eXchange Mice via Pronuclear Transfer
Published: Vol 6, Iss 20, Oct 20, 2016 DOI: 10.21769/BioProtoc.1976 Views: 12338
Reviewed by: Masahiro MoritaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Conditional Human BRD4 Knock-In Transgenic Mouse Genotyping and Protein Isoform Detection
Michael Paul Lewis [...] Cheng-Ming Chiang
Apr 5, 2022 3618 Views

Arrayed CRISPR/Cas9 Screening for the Functional Validation of Cancer Genetic Dependencies
Ludovica Proietti [...] Florian Grebien
Dec 20, 2022 3082 Views

Leveraging Circular Polymerization and Extension Cloning (CPEC) Method for Construction of CRISPR Screening Libraries
Bengisu Dayanc [...] Serif Senturk
Feb 20, 2025 2775 Views
Abstract
The mitochondrial paradigm for common disease proposes that mitochondrial DNA (mtDNA) sequence variation can contribute to disease susceptibility and progression. To test this concept, we developed the Mitochondrial-nuclear eXchange (MNX) model, in which isolated embryonic pronuclei from one strain of species are implanted into an enucleated embryo of a different strain of the same species (e.g., C57BL/6 and C3H/HeN, Mus musculus), generating a re-constructed zygote harboring nuclear and mitochondrial genomes from different strains. Two-cell embryos are transferred to the ostia of oviducts in CD-1 pseudopregnant mice and developed to term. Nuclear genotype and mtDNA haplotype are verified in offspring, and females selected as founders for desired MNX colonies. By utilizing MNX models, many new avenues for the in vivo study for mitochondrial and nuclear genetics, or mito-Mendelian genetics, are now possible.
Keywords: MitochondriaBackground
The isolation of nuclear and mitochondrial genomes in MNX mice strains allows examination of pathomechanisms of dysfunctional bioenergetics such as cardiovascular disease (Fetterman et al., 2013; Grimsditch et al., 2000; Paigen et al., 1990; Wang et al., 2005), glucose tolerance (Freeman et al., 2006; Kaku et al., 1988) and fatty liver disease (Betancourt et al., 2014). This approach is distinct from conplastic (Yu et al., 2009) and xenomitochondrial (McKenzie et al., 2004) approaches in that MNX mice are generated directly with 100% of the desired nuclear and mtDNA complements from respective donor strains through nuclear transfer and thus do not require repeated back-crossings (as do conplastics) to generate animals having the desired genotype (Figure 1). Furthermore, MNX mice allow direct, unambiguous assessment of mtDNA contributions to disease since there is no complexity introduced by potential nuclear cross-over and combinational effects in the filial generations associated with standard backcrossing methods used to generate conplastic mice.
Materials and Reagents
- 26 gauge needle
- 1 or 3 ml syringe
- Microscope slides
- CellTram vario syringe
- 10 centimeter tissue culture dishes
- Female donor mice (3-4 weeks of age) of desired nuclear or mitochondrial genetic backgrounds
- Male breeders (proven) of matching nuclear background as donor females
- Female recipient mice (8-10 weeks of age)
- Vasectomized male mice (proven)
- Gonadotropin from pregnant mare serum (PMS) (Sigma-Aldrich, catalog number: G4877 )
Note: This product has been discontinued. - Human chorionic gonadotropin (HCG) (Sigma-Aldrich, catalog number: CG10 )
- M2 medium (Sigma-Aldrich, catalog number: M7167 )
- Cytochalasin B from Drechslera dematioidea (Sigma-Aldrich, catalog number: C6762 )
- Colcemid (Sigma-Aldrich, catalog number: D1925 )
- Embryo tested mineral oil (Sigma-Aldrich, catalog number: M8410 )
- Water for embryo transfer (Sigma-Aldrich, catalog number: W1503 )
- Restriction enzyme:
- BclI (New England Biolabs)
- PflFI/AspI (New England Biolabs)
- Dilution of PMS (see Recipes)
- Dilution of HCG (see Recipes)
- Dilution of cytochalasin B (see Recipes)
- Dilution of colcemid (see Recipes)
Equipment
- Piezoelectric drill (piezo drill) (Sutter Instrument, model: PrimeTech PMM-150FU )
- Electroporator (BTX The Electroporation Experts, model: ECM 630 )
- Micropipette puller (horizontal pipette puller) (Sutter Instrument, model: P-87 )
The product P-87 has been discontinued and the available one is P-97 . - Micropipette microforge (Defonbrune microforge with microscope head) (Leitz)
- Holding pipette puller (vertical pipette puller) (David Kopf Instrument, model: 720 )
- Microscope (Laborlux S Nomarski DIC) (Leica Microsystems)
- CellTram Vario syringe (Eppendorf)
- Benchtop incubator (Cook, model: MINC G20079 )
- 45 degree angled forceps (Fine Science Tools, catalog number: 11251-35 )
- Straight forceps (Fine Science Tools, catalog number: 11251-10 )
- Microdissecting spring scissors (Roboz Surgical Instrument, catalog number: RS-5650 )
- Mouth pipette
Procedure
- Creation of MNX mice
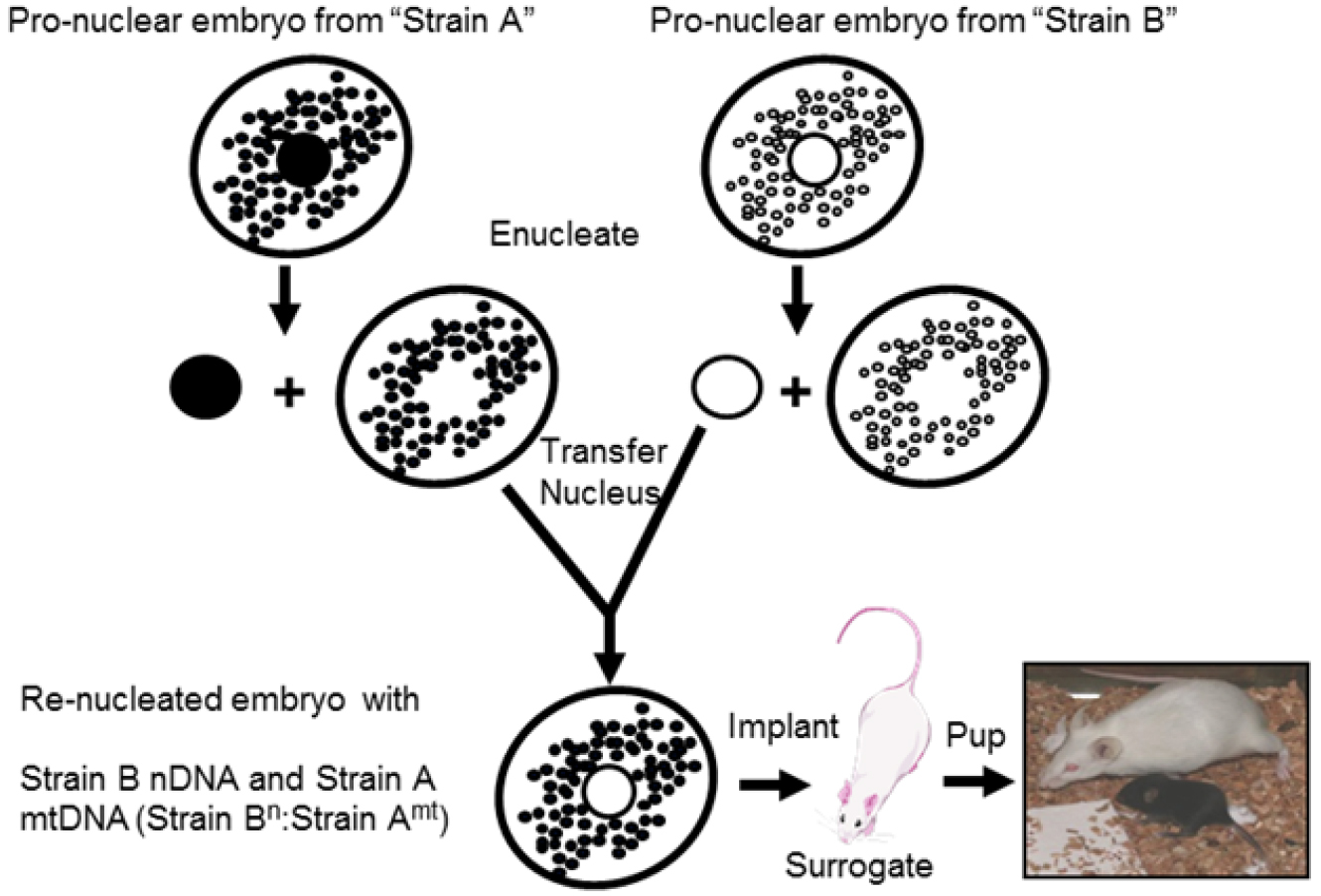
Figure 1. Overall procedure schematic. MNX mice were produced by enucleating fertilized oocytes from 'Strain A' (C57BL/6) and 'Strain B' (C3H/HeN) mice. MNX nomenclature is indicated by strain nuclear (Strainn):strain mtDNA (Strainmt), e.g., mice with C3H nuclear DNA and C57 mtDNA are indicated by C3Hn:C57mt. Strain B pronuclei were transferred to enucleated Strain A oocytes yielding Strain Bn:Strain Amt (C3Hn:C57mt) oocytes, which were implanted into surrogate females to generate MNX progeny. The reciprocal process was followed to generate the Strain An:Strain Bmt (C57n:C3Hmt) oocytes.- Female donor mice are injected intraperitoneally (IP) with PMS (5.0-7.5 IU, in a volume 0.2-0.3 ml) using a 26 gauge needle and a 1 or 3 ml syringe. Optimum dose and age of the mice can be strain specific, and should be determined via dose response curves with assessment of fertilized embryo production. Typically, female mice are injected ~noon on day 1.
- Approximately 48 h after (day 3) the PMS injection, donor mice receive an IP injection of HCG of the same dose as PMS. Immediately following injection, super-ovulated females are paired and mated with proven breeder males of the same nuclear genotype (matching the female, Figure 1); often copulation occurs within the first 1-2 h of mating. Female recipient mice positive for signs of proestrus by visual inspection are paired with proven vasectomized males to generate pseudopregnancy (Byers et al., 2012).
- Females are checked the following morning for vaginal plugs. The presence of a plug indicates successful mating, but does not necessarily indicate fertilization. The absence of a plug does not however confirm that successful breeding did not occur since facile identification of plugs can be strain dependent.
- Cumulus masses are harvested as previously described to produce single-celled embryos (Figures 2A and 2B) (Han et al., 2010).
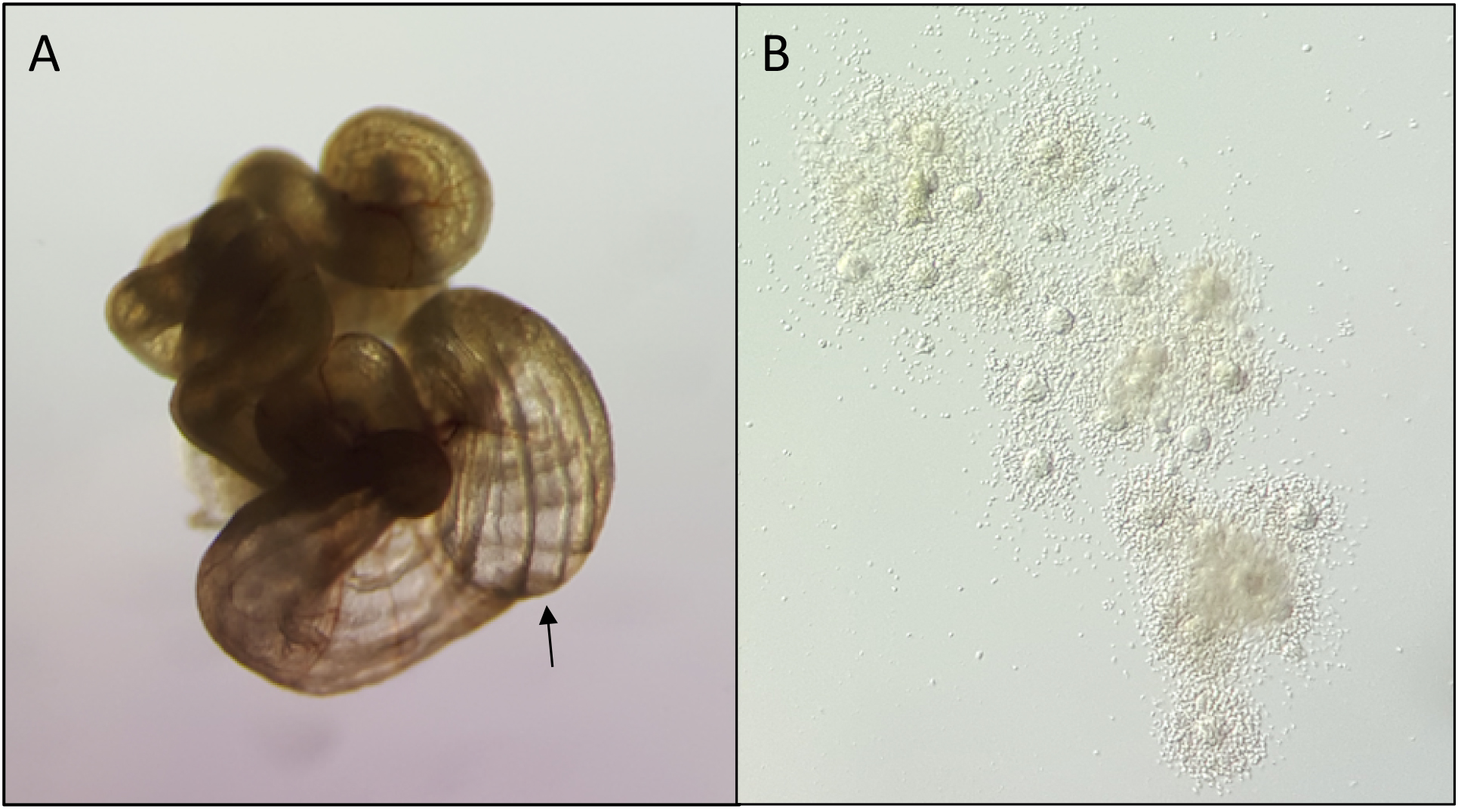
Figure 2. Collection of cumulus masses from oviducts. Oviducts (A) are dissected from superovulated females with vaginal plugs. The swollen ampulla (indicated by arrow) is nicked to allow the embryos and cumulus cells to be expelled (B). - Pronuclear embryos are placed by mouth pipette in M2 medium containing cytochalasin B (5 μg/ml) and colcemid (0.1 μg/ml) at 37 °C for 5 min, and remain in a microinjection drop of M2 medium on a microscope slide at room temperature to prevent lysis during manipulation.
- A micropipette similar in size and shape to a beveled pipet used for embryonic stem cell injections (approximately 20 micron inner diameter) is used to remove both pronuclei of each embryo by applying slight pressure on the zona pellucida of the first embryo (Longenecker and Kulkarni, 2009). A high-intensity piezo pulse is applied only until the zona is ruptured (approximately 1-2 sec) and then turned off. The pipet is slowly advanced to each pronucleus (Figure 3), and with gentle suction applied to the needle using a CellTram vario syringe, the two pronuclei are aspirated and removed as a single unit (karyoplast, Figure 3). Appropriate needle size and application of suction only when the pronuclei are in contact with the needle minimizes uptake of cytoplasmic contents into the needle.
- The procedure is repeated on the reciprocal embryo, resulting in the pipet containing pronuclei from both strains (Figure 3).
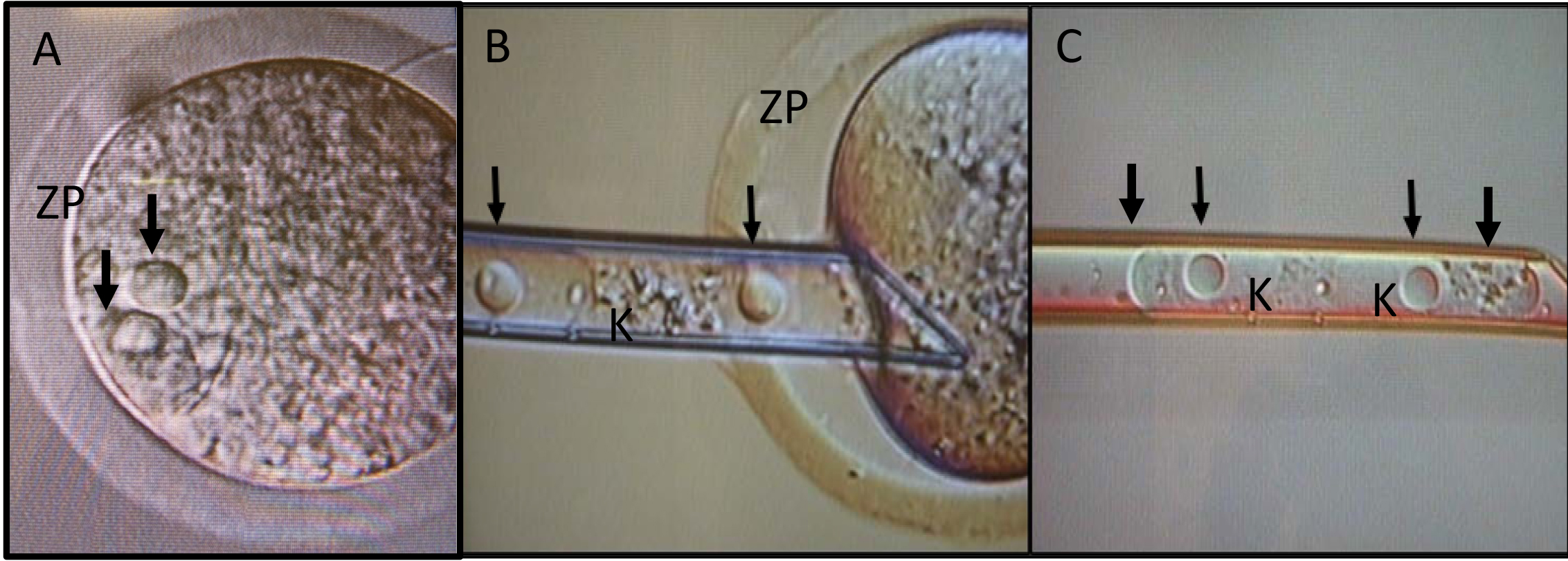
Figure 3. Isolation of pronuclei. (A) The pipet is slowly advanced to each pronucleus (indicated by black arrows), and with gentle suction applied to the needle using a CellTram vario syringe (B) the two pronuclei are aspirated and removed (C) as a single unit (karyoplast, K). ZP, zona pellucida. - The isolated pronuclei from Strain A are implanted into the enucleated embryo of Strain B (Figure 4). The reciprocal karyoplasts from Strain B are placed into the enucleated Strain A embryos.
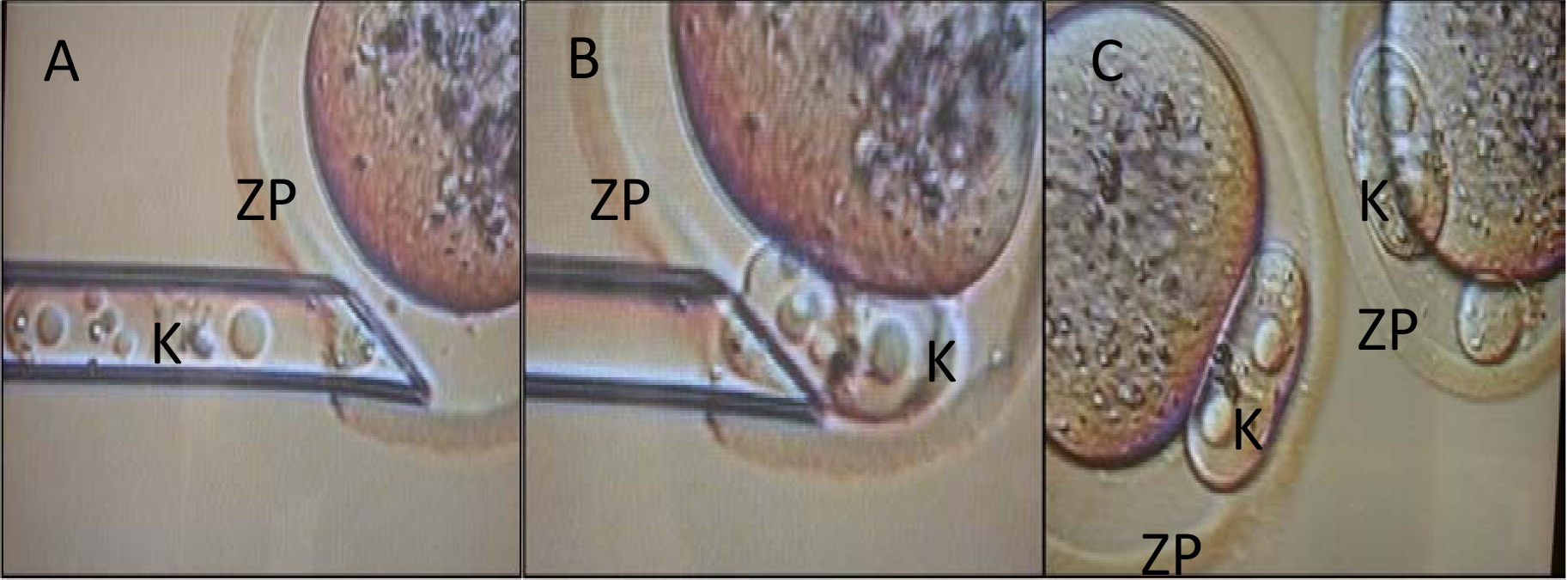
Figure 4. Transfer of pronuclei. The karyoplasts (K) from one strain are then implanted into the enucleated embryos of the reciprocal strain. ZP, zona pellucida. (A) Insertion of pipet containing karyoplasts (B) Ejection of karyoplasts (C) Reconstructed embryo. - Ten centimeter tissue culture dishes (Figure 5) are loaded with 30 μl microdrops of M2 medium and covered in mineral oil and each embryo is placed by mouth pipette into its own drop of media. Strain A and Strain B embryos are placed in separate dishes. An electrode is placed into the drop positioning the embryo between the two poles. A single 90 V pulse is applied to each re-constructed zygote and all zygotes are cultured overnight (Han et al., 2010).
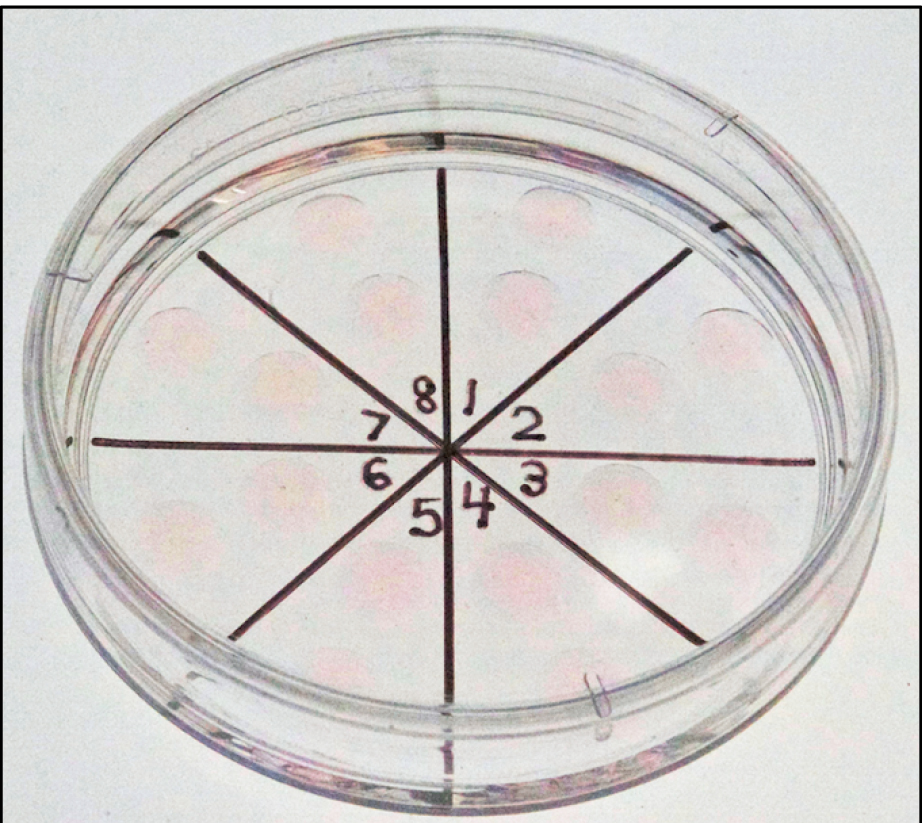
Figure 5. Embryo culture dish. Panels 1-8 are different drops of M2 medium and mineral oil containing reconstructed zygotes. Each drop may contain 1-2 zygotes. - Two-cell embryos are transferred to the ostia of oviducts of 0.5 day pseudopregnant mice to term; approximately 20 embryos are transferred per recipient (Figure 6).
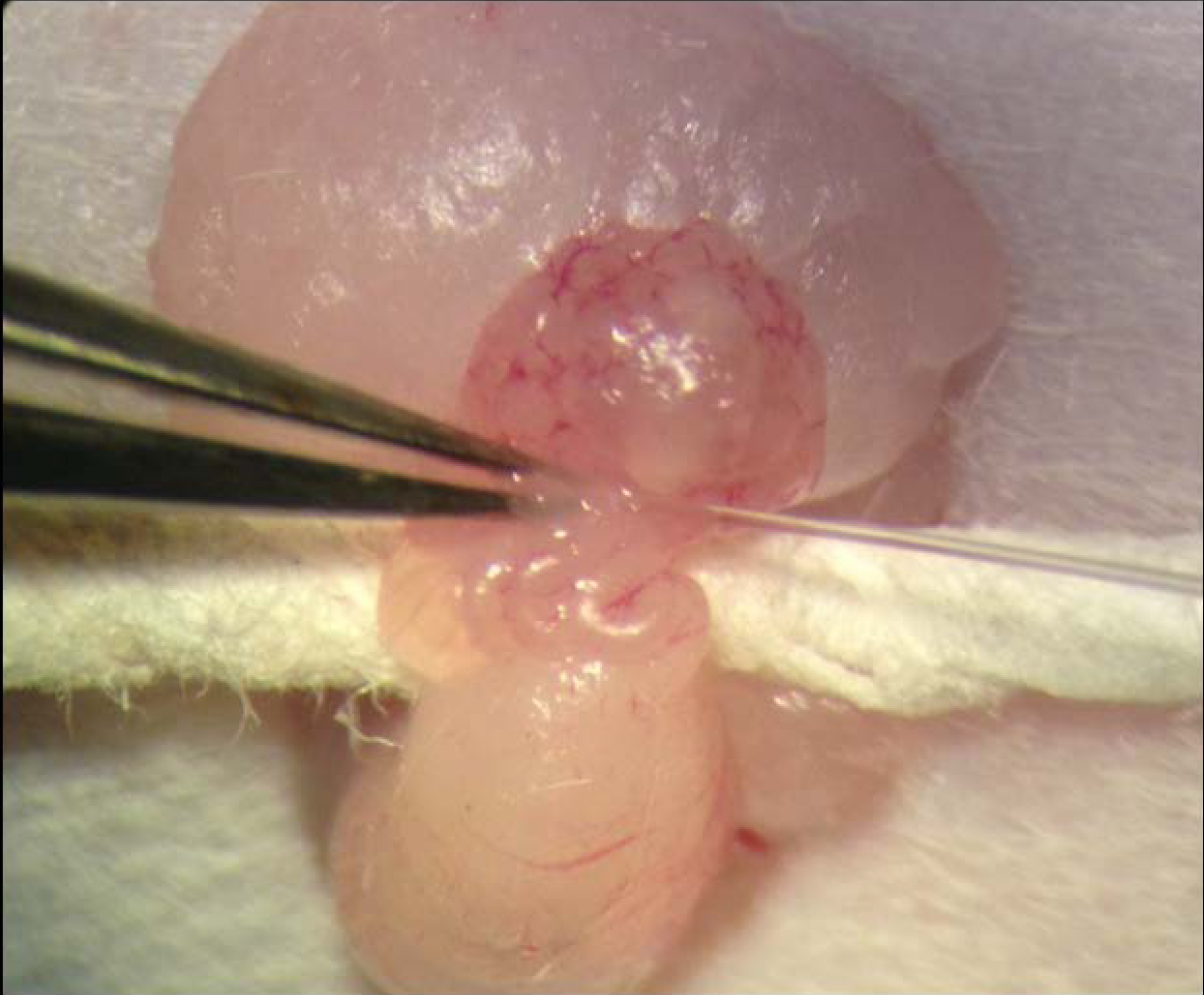
Figure 6. Oviduct transfer of reconstituted embryos. Two cell embryos are placed into the oviduct of a pseudopregnant recipient mouse through the infundibular ostium by mouth pipet. - Nuclear genotype and mtDNA haplotype are verified in offspring via nuclear SNP and complete mtDNA genome sequence analysis from ear or tail clips.
- Female donor mice are injected intraperitoneally (IP) with PMS (5.0-7.5 IU, in a volume 0.2-0.3 ml) using a 26 gauge needle and a 1 or 3 ml syringe. Optimum dose and age of the mice can be strain specific, and should be determined via dose response curves with assessment of fertilized embryo production. Typically, female mice are injected ~noon on day 1.
- MNX mouse colony establishment
MNX mouse colonies are established using homoplasmic founder female MNX mice based upon PCR analysis specific for each potential mtDNA haplotype. mtDNA haplotype is initially confirmed by PCR detection methods developed specifically for each haplotype and homoplasmic females confirmed by sequencing. Desired homoplasmic (based upon PCR screening) females to be used as founders are identified in ~20%-30% of generated females.- MNX females of verified mtDNA sequence and apparent homoplasmy (using ear or tail clip DNA) are crossed with males of the matching nuclear background to generate F1 litters.
- Multiple tissues from F1 progeny are tested to verify desired mtDNA haplotype/homoplasmy via PCR analysis designed specifically for diagnostic mtDNA SNP’s (Bayona-Bafaluy et al., 2003). For example, F1 C57n:C3Hmt and C3Hn:C57mt mice were haplogrouped in multiple tissues based upon a PCR detection method to interrogate the potential for heteroplasmy (Figure 7).
PCR protocol:
2x GoTaq MasterMix: 25 μl
Primers: 5 μl each
Nuclease free H2O: 12.5 μl
DNA: 2.5 μl
Thermocycling profile:
95 °C for 2 min: 1 cycle
95 °C for 30 sec; 57 °C for 1min; 72 °C for 30 sec: 35 cycles
72 °C for 10 min: 1 cycle
24 °C for infinity- To screen for the ND3 mutation at bp 9461 (204 base pair amplicon):
9461F: 5’-TTCCAATTAGTAGATTCTGAATAAACCCAGAAGAGAGTGAT-3’
9461R: 5’-AAATTTTATTGAGAATGGTAGACG-3’
Ten microliters of amplicon are digested with the restriction enzyme Bcli (10 U, New England Biolabs) in a 10 microliter reaction volume. The C3H mtDNA is cleaved into 166 bp and 38 bp fragments, while the C57 mtDNA remains uncut. - To screen for the bp 9348 CO3 mutation (385 base pair amplicon):
9348F: 5’- CGAAACCACATAAATCAAGCCC-3’
9348R: 5’-CTCTCTTCTGGGTTTATTCAGA-3’
Ten microliters of amplicon are digested using the Pflf1 (AspI, 10 U, New England Biolabs) restriction enzyme in a 20 microliter reaction volume. The C3H mtDNA remains uncut while C57 mtDNAs are digested into 274 bp and 111 bp fragments.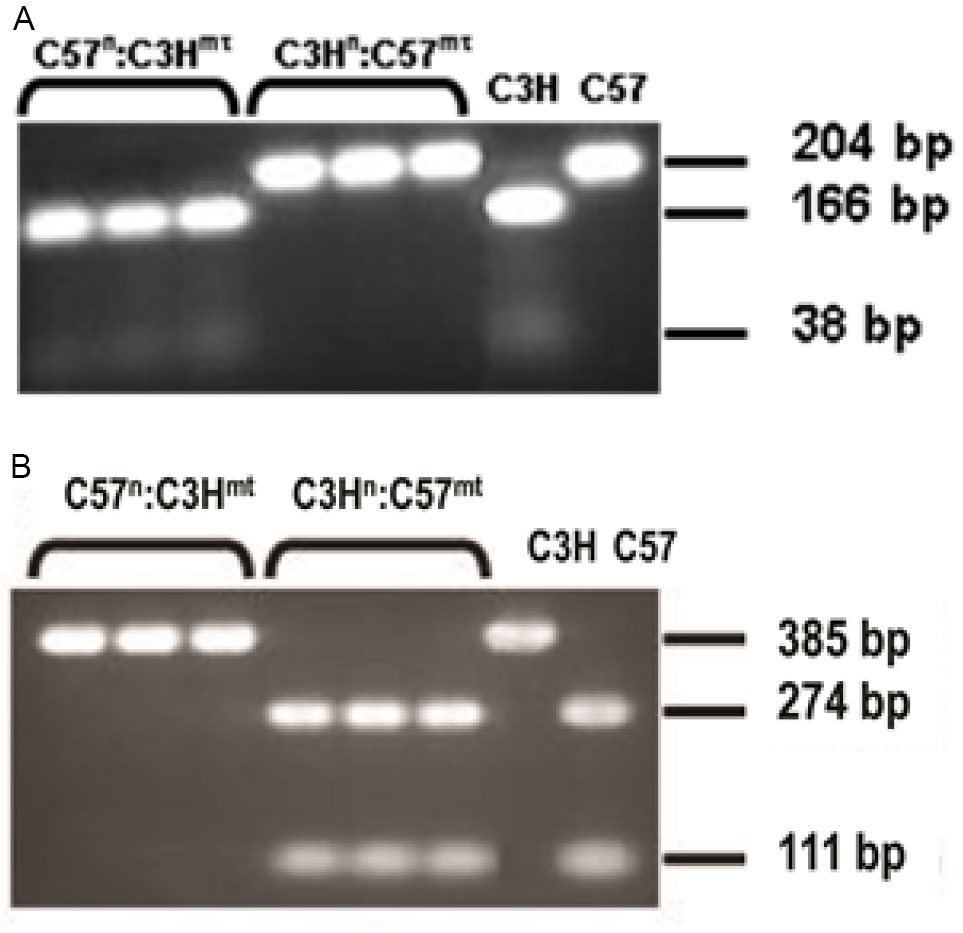
Figure 7. Generation of Mitochondrial-nuclear eXchange (MNX) mice. Nuclear genotyping and mtDNA haplotyping of all originating founding females and F1 progeny was determined by nuclear SNP analysis of a panel of 38 distinguishing nuclear SNPs and complete sequencing of the mtDNA. After initial mtDNA haplotype verification by direct sequencing for founders, subsequent generations are haplotyped via restriction enzyme length polymorphism analysis using AspI and BclI which give patterns distinct for C3H and C57 mtDNAs. A. PCR products from C57 mtDNAs remain uncut (204 bp) whereas C3H mtDNAs are cleaved by BclI to yield 166 bp and 38 bp fragments. B. PCR products from C57 mtDNAs are cleaved by AspI to yield 274 bp and 111 bp fragments whereas C3H mtDNAs remain uncut (385 bp).
- To screen for the ND3 mutation at bp 9461 (204 base pair amplicon):
- MNX female founders that produced mice which appear homoplasmic in all tested tissues are then used as founding dams for each respective MNX colony.
- All progeny of each generation are mtDNA haplogrouped using PCR based techniques to verify maintenance of desired genetic background.
- MNX females of verified mtDNA sequence and apparent homoplasmy (using ear or tail clip DNA) are crossed with males of the matching nuclear background to generate F1 litters.
Data analysis
The original research paper detailing generation of MNX mice as well as analysis and replicate information is available online (Fetterman et al., 2013).
Recipes
- Dilution of PMS
Add 20 ml embryo tested water to 1,000 IU bottle of lyophilized PMS to create 20 ml stocks at 50 IU/ml.
Store at -20 °C for up to 3 months. - Dilution of HCG
Add 10 ml embryo tested water to 10,000 IU bottle of lyophilized HCG to create 10 stocks at 1,000 IU.
Dilute each tube as needed with 20 ml embryo tested water to create 20 ml stocks at 50 IU/ml.
Store at -20 °C for up to 6 weeks. - Dilution of cytochalasin B
Add 1 ml DMSO to 1 mg cytochalasin B powder to create 1 mg/ml stock solution.
Dilute 5 μl stock in 1 ml M2 medium for a 5 μg/ml solution.
Store at -20 °C for up to 6 months. - Dilution of colcemid
Add 10 μl of purchased, 10 μl/ml colcemid stock solution, and 1 ml M2 medium to create a 0.1 μg/ml solution.
Store at -20 °C for up to 6 months.
Acknowledgments
This work was funded by the National Institutes for Health (grant numbers RO1 94518 and RO1103859 [to S.W.B.]), the National Foundation for Cancer Research (to D.R.W.), Susan G. Komen for the Cure (grant number SAC111370 [to D.R.W.]), U.S. Army Medical Research & Material Command (grant number W81XWH-07-1-0540d [to S.W.B.]) and a pilot grant from the University of Alabama at Birmingham Comprehensive Cancer Center (grant number CA013148 [to D.R.W. and S.W.B.]). Additional support was received from the National Institutes of Health (grant number RO1 HL109785 [to L.J.D.]), a National Institutes of Health predoctoral training programme in cardiovascular pathophysiology (grant number T32 HL007918 [to J.L.F.]), American Heart Association predoctoral fellowships (grant numbers 09PRE2240046 [to J.L.F.] and 11PRE7650033 [to K.J.D.]). The University of Alabama at Birmingham Transgenic Mouse Facility (RAK) is supported by the National Institutes of Health [grant numbers P30 CA13148, P30 AR048311, P30 DK074038, P30 DK05336 and P60 DK079626] and by the National Institutes of Health-funded Diabetes Research Center Bioanalytical Redox Biology Core [grant number P60 DK079626] located at the University of Alabama at Birmingham.
References
- Bayona-Bafaluy, M. P., Acin-Perez, R., Mullikin, J. C., Park, J. S., Moreno-Loshuertos, R., Hu, P., Perez-Martos, A., Fernandez-Silva, P., Bai, Y. and Enriquez, J. A. (2003). Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res 31(18): 5349-5355.
- Betancourt, A. M., King, A. L., Fetterman, J. L., Millender-Swain, T., Finley, R. D., Oliva, C. R., Crowe, D. R., Ballinger, S. W. and Bailey, S. M. (2014). Mitochondrial-nuclear genome interactions in non-alcoholic fatty liver disease in mice. Biochem J 461(2): 223-232.
- Byers, S. L., Wiles, M. V., Dunn, S. L. and Taft, R. A. (2012). Mouse estrous cycle identification tool and images. PLoS One 7(4): e35538.
- Fetterman, J. L., Zelickson, B. R., Johnson, L. W., Moellering, D. R., Westbrook, D. G., Pompilius, M., Sammy, M. J., Johnson, M., Dunham-Snary, K. J., Cao, X., Bradley, W. E., Zhang, J., Wei, C. C., Chacko, B., Schurr, T. G., Kesterson, R. A., Dell'italia, L. J., Darley-Usmar, V. M., Welch, D. R. and Ballinger, S. W. (2013). Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload. Biochem J 455(2): 157-167.
- Freeman, H. C., Hugill, A., Dear, N. T., Ashcroft, F. M. and Cox, R. D. (2006). Deletion of nicotinamide nucleotide transhydrogenase: a new quantitive trait locus accounting for glucose intolerance in C57Bl/6J mice. Diabetes 55(7): 2153-2156.
- Grimsditch, D. C., Penfold, S., Latcham, J., Vidgeonhart, M., Groot, P. H. and Benson, G. M. (2000). C3H apoE(-/-) mice have less atherosclerosis than C57BL apoE(-/-) mice despite having a more atherogenic serum lipid profile. Atherosclerosis 151(2): 389-397.
- Han, Z., Cheng, Y., Liang, C. G. and Latham, K. E. (2010). Nuclear transfer in mouse oocytes and embryos. Methods Enzymol 476(2): 171-84.
- Kaku, K., Jr, F. F., Province, M. and Permutt, M. A. (1988). Evidence for polygenic control. Diabetes 37(6): 707-13.
- Longenecker, G. and Kulkarni, A. B. (2009). Generation of gene knockout mice by ES cell microinjection. Curr Protoc Cell Chapter 19, Unit 19.14 19.14.1-36.
- Mckenzie, M., Trounce, I. A., Cassar, C. A., and Pinkert, C. A. (2004). Production of homoplasmic xenomitochondrial mice. Proc Natl Acad Sci U S A 101(6): 1685-1690.
- Paigen, B., Morrow, A., Brandon, C., Mitchell, D. and Holmes, P. (1990). Variation in susceptibility to atherosclerosis among inbred strains of mice. Atherosclerosis 10(2): 65-73.
- Wang, X., Ria, M., Kelmenson, P.M., Eriksson, P., Higgins, D.C., Samnegård, A., Petros, C., Rollins, J., Bennet, A.M., Wiman, B., de Faire, U., Wennberg, C., Olsson, P.G., Ishii, N., Sugamura, K., Hamsten, A., Forsman-Semb, K., Lagercrantz, J. and Paigen, B. (2005). Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat Genet 37(4): 365-372.
- Yu, X., Gimsa, U., Wester-Rosenlöf, L., Kanitz, E., Otten, W., Kunz, M. and Ibrahim S. M. (2009). Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Res 19(1): 159-165.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kesterson, R. A., Johnson, L. W., Lambert, L. J., Vivian, J. L., Welch, D. R. and Ballinger, S. W. (2016). Generation of Mitochondrial-nuclear eXchange Mice via Pronuclear Transfer. Bio-protocol 6(20): e1976. DOI: 10.21769/BioProtoc.1976.
Category
Molecular Biology > DNA > Mutagenesis
Molecular Biology > DNA > Genotyping
Molecular Biology > DNA > DNA cloning
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











