- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Colorimetric Method to Measure Plant Glutamate Dehydrogenase Enzyme Activity
Published: Vol 5, Iss 16, Aug 20, 2015 DOI: 10.21769/BioProtoc.1560 Views: 12058
Reviewed by: Arsalan DaudiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1465 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3196 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1811 Views
Abstract
Glutamate dehydrogenase (GDH) is an NAD(H) dependent enzyme that catalyzes, in vitro, the reversible amination of glutamate. Here we describe how to determine spectrophotometrically GDH activity monitoring NADH evolution. This protocol is described here for Arabidopsis thaliana (A. thaliana) although it is also valid for other plant species. GDH protein is a hexamer composed, in the case of Arabidopsis, of a combination of GDHα, GDHβ and GDHγ subunits. Every combination of subunits is possible; however, it is still barely known whether different combinations affect the enzymatic properties of the hexamers. In other species, hexamers are a combination of GDHα and GDHβ but it cannot be discarded the existence of other genes since for instance GDHγ subunit in Arabidopsis was described in Fontaine et al. (2012).
Glutamate + NAD+ + H+ → 2-Oxoglutarate + NADH + NH4+
Materials and Reagents
- 4-weak old Arabidopsis thaliana leaves and roots
- Ultrapure water
- Liquid N2
- Ice
- Flat bottom microplates (Deltalab, catalog number: 900011.1 )
- 4 mm diameter glass beads (Glaswarenfabrik Karl Hecht) (VWR International, catalog number: 201-0278 )
- 1.2 ml deep well storage plate (Thermo Fisher Scientific, catalog number: AB-0564 )
- EVA Capband for capping 8 tubes (Micronic, catalog number: MP227B1 )
- Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories, AbD Serotec®, catalog number: 500-006 )
- NADH (Sigma-Aldrich, catalog number: 43420 )
- PVPP (Sigma-Aldrich, catalog number: 77627 )
- Glycerol (EMD Millipore Corporation, catalog number: 104092 )
- Bovine Serum Albumin (BSA) (Sigma-Aldrich, catalog number: A7906 )
- Triton X-100 (Sigma-Aldrich, catalog number: T-9284 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- KOH (EMD Millipore Corporation, catalog number: 105029 )
- MnCl2 (Sigma-Aldrich, catalog number: M8266 )
- EDTA (Sigma-Aldrich, catalog number: ED255 )
- EGTA (Sigma-Aldrich, catalog number: E4378 )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632 )
- PMSF (Sigma-Aldrich, catalog number: P7626 )
- Delta-aminocaproic acid (Sigma-Aldrich, catalog number: A2504 )
- Leupeptin (Sigma-Aldrich, catalog number: L2884 )
- Tricine (Sigma-Aldrich, catalog number: T0377 )
- CaCl2 (Panreac Applichem, catalog number: 131232 )
- 2-Oxogutarate (Merck KGaA, catalog number: 5194 )
- (NH4)2SO4 (Panreac Applichem, catalog number: 131140 )
- Extraction buffer (see Recipes)
- Reaction buffer (see Recipes)
- Bradford solution (see Recipes)
Equipment
- TissueLyser (RETSCH, model: MM400 )
- Plate centrifuge (Sigma, model: 2-16K )
- Absorbance microplate reader (Biotek Powerwave X 384 Microplate spectrophotometer)
- TissueLyser Adapter Set 2 x 96 (QIAGEN, catalog number: 69984 )
Procedure
- Material harvest and homogenization
- In a 96 deepwell (1.2 ml) plate, place 2 glass beads (4 mm diameter) into each well.
- Weigh approximately 20 mg of fresh leaves or roots and place them in a 96-deepwell plate, half-submerged in liquid nitrogen. Store sealed with EVA Capband at -80 °C until use.
- Grind frozen tissue two times with a TissueLyser for 60 sec at 27 Hz frequency. Plates are coupled to the TissueLyser with a TissueLyser Adapter Set 2 x 96. (Ensure sealing of the wells during homogenization).
- In a 96 deepwell (1.2 ml) plate, place 2 glass beads (4 mm diameter) into each well.
- Extraction
- Add around 3-5 mg PVPP per well (this approximately corresponds to a level micro spatula).
- Add 500 µl of cold extraction buffer to each well and rapidly mix by inverting the plate (avoid thawing the samples prior to adding the buffer).
- Homogenize the samples again with TissueLyser two times for 60 sec at 27 Hz frequency.
- Centrifuge the plate at 4,000 rpm and 4 °C for 30 min in a plate centrifuge.
- Recover the supernatants (400 µl) in a new 96 deepwell plate and keep the samples on ice.
- Add around 3-5 mg PVPP per well (this approximately corresponds to a level micro spatula).
- Determination of protein content by Bradford method
- In a 96-well spectrophotometer plate, place 1 µl of extract per well and add ultrapure water up to 10 µl. Make a blank with 1 µl of extraction buffer.
- For the calibration curve place BSA standards in a range of 0 to 6 µg of BSA/well using a 1 µg/µl BSA stock and complete volume up to 10 µl.
- Add in every well 240 µl of Bradford solution and incubate for 5 min at room temperature in the dark.
- Read absorbance at 595 nm.
- In a 96-well spectrophotometer plate, place 1 µl of extract per well and add ultrapure water up to 10 µl. Make a blank with 1 µl of extraction buffer.
- Enzyme activity measurement
- Place 20 µl of supernatant per well.
- Add 280 µl of reaction buffer. This step should be performed rapidly since the reaction starts as soon as the buffer is added. Thus, if measuring 96 samples the use of a multichannel pipette is strongly advised to ensure reproducibility.
- Immediately monitor NADH evolution kinetic at 340 nm in the spectrophotometer for 10 min at 30 °C (Figure 1). Ensure spectrophotometer is already at 30 °C before adding the reaction buffer to the sample.
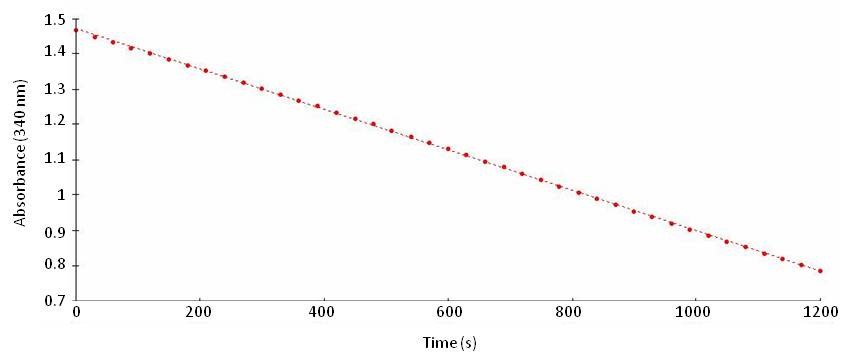
Figure 1. Graph showing NADH consumption by GDH aminating activity
- For NADH calibration curve, place 300 µl NADH dilutions from 0-0.2 mM in a microplate and read at 340 nm. (NADH dilutions must be prepared just prior to the measurement of the calibration curve to avoid the oxidation of NADH).
- Place 20 µl of supernatant per well.
Data analysis
- To determine protein concentration in the extracts, subtract from the absorbance of every sample the absorbance of the blank (done with extraction buffer) and calculate the protein concentration with the calibration curve performed with BSA standards.
- Enzyme activity is calculated with the maximum speed (Vmax) obtained from NADH oxidation monitoring at 340 nm (don’t take the two first minutes of the measurement into account and ensure linearity of the activity). Subtract the blank (made with extraction buffer) from every sample and calculate GDH activity with NADH calibration curve. Finally express the activity per µmol of NADH consumed per minute and per µg of protein (obtained from Bradford assay) or per mg of fresh/dry weight.
Notes
- This protocol can be easily adapted to eppendorf tubes and spectrophotometer cuvettes instead of 96-well plates just scaling the volumes. Similarly the homogenization of the samples might be done with a mortar and pestle instead of the TissueLyser.
- If possible, we advice to do every 96-well plate step using multi-channel pipettes to save time and thus to gain reproducibility among samples.
- This protocol is for Arabidopsis thaliana but it can be used for every species. For some species desalting of protein extracts supernatants might be considered.
Recipes
Note: All solutions should be made in ultrapure water.
- Extraction buffer
Glycerol 10%
BSA 0.05%
Triton X-100 0.1%
50 mM HEPES (pH 7.5, adjusted with KOH)
10 mM MgCl2
1 mM EDTA
1 mM EGTA
1 mM PMSF
10 mM DTT
1 mM Delta-aminocaproic acid
1 µM Leupeptin
- Reaction buffer (pH 8)
100 mM Tricine (pH 8, adjusted with KOH)
1 mM CaCl2
50 mM (NH4)2SO4
0.25 mM NADH
13 mM 2-Oxoglutarate
- Bradford solution
Dilute Bio-Rad Protein Assay Dye Reagent Concentrate 1:5 with ultrapure water
Stored in a dark bottle at 4 °C
Acknowledgments
This protocol is adapted from Sarasketa et al. (2014) and based on the methodology reported by Groat and Vance (1981). This work was supported by the Basque Government (IT526-10), the UPV/EHU (EHUA14/14), the People Program (Marie Curie Actions) of the European Union’s Seventh Framework Program (FP7/2007–2013) under REA grant agreement number 334019 and MINECO (BIO2014-56271-R).
References
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Fontaine, J. X., Terce-Laforgue, T., Armengaud, P., Clement, G., Renou, J. P., Pelletier, S., Catterou, M., Azzopardi, M., Gibon, Y., Lea, P. J., Hirel, B. and Dubois, F. (2012). Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 24(10): 4044-4065.
- Groat, R. G. and Vance, C. P. (1981). Root nodule enzymes of ammonia assimilation in alfalfa (Medicago sativa L.): DEVELOPMENTAL PATTERNS AND RESPONSE TO APPLIED NITROGEN. Plant Physiol 67(6): 1198-1203.
- Sarasketa, A., Gonzalez-Moro, M. B., Gonzalez-Murua, C. and Marino, D. (2014). Exploring ammonium tolerance in a large panel of Arabidopsis thaliana natural accessions. J Exp Bot 65(20): 6023-6033.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Sarasketa, A., Vega-Mas, I. and Marino, D. (2015). In vitro Colorimetric Method to Measure Plant Glutamate Dehydrogenase Enzyme Activity. Bio-protocol 5(16): e1560. DOI: 10.21769/BioProtoc.1560.
Category
Plant Science > Plant biochemistry > Protein > Activity
Plant Science > Plant metabolism > Nitrogen
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









