- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Circular RT-PCR Assay Using Arabidopsis Samples
Published: Vol 5, Iss 14, Jul 20, 2015 DOI: 10.21769/BioProtoc.1533 Views: 17544
Reviewed by: Arsalan DaudiYuan ChenAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Real-time PCR Analysis of PAMP-induced Marker Gene Expression in Nicotiana benthamiana
Fan Liu [...] Yuanchao Wang
Oct 5, 2018 9304 Views

RNA Stability Measurements Using RT-qPCR in Arabidopsis Seedlings
Tianran Jia and Brandon H. Le
Jul 20, 2020 6996 Views

Laser-Assisted Microdissection and High-Throughput RNA Sequencing of the Arabidopsis Gynoecium Medial and Lateral Domains
Valentín Luna-García and Stefan de Folter
Sep 5, 2024 2066 Views
Abstract
Post-transcriptional processing is critical for RNA biogenesis, in which conventional functional RNA transcripts are generated, such as messenger RNAs (mRNAs), transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs) for translation as well as emerging non-coding RNAs with known or unknown regulatory functions. To determine the precise termini of an RNA molecule during or after processing, the primer extension and Rapid Amplification of cDNA Ends (RACE) methods have been routinely utilized for the precise mapping of 5’ or 3’ ends. Different from these assays, which are designed to detect only one end of a specific target RNA at a time, circular Reverse Transcription-Polymerase Chain Reaction (cRT-PCR) is able to simultaneously determine both the 5’ and 3’ ends of the target RNA. In Arabidopsis thaliana, cRT-PCR has been wildly applied to identify both the 5’ and 3’ extremities of the ribosomal RNA precursors, or to assess the length or post-transcriptional extensions at the 3’ end of a matured mRNA. In this protocol, we summarize and present a detailed procedure of the cRT-PCR assay in Arabidopsis thaliana, which is also successfully used in our previously published work (Hang et al., 2014).
Keywords: Circular RT-PCRMaterials and Reagents
- Arabidopsis thaliana 14-day-old seedlings
- M13F: 5′-TGTAAAACGACGGCCAGT-3′ and M13R: 5’-CAGGAAACAGCTATGAC-3’
- TRNzol Reagent (TIANGEN®, catalog number: DP40502 )
- Dynabeads® mRNA Purification Kit (Life Technologies, catalog number: 61006 )
- FirstChoice® RLM-RACE Kit (Life Technologies, catalog number: AM1700 )
- RNA 5’ Polyphosphatase (Epicentre, catalog number: RP8092H )
- T4 RNA Ligase 1 (New England Biolabs, catalog number: M0204S )
- T4 RNA Ligase Reaction Buffer (New England Biolabs, catalog number: B0216L )
- Adenosine 5'-Triphosphate (New England Biolabs, catalog number: P0756S )
- RNasin® Plus RNase Inhibitor (Promega Corporation, catalog number: N2615 )
- Glycogen (RNA grade) (Thermo Fisher Scientific, catalog number: R0551 )
- TransScript® II First-Strand cDNA Synthesis SuperMix (TransGen Biotech, catalog number: AH301 )
- TaKaRa ExTaq® DNA Polymerase (Takara Bio Company, catalog number: RR001B )
- pEASY® -T1 Cloning Kit (TransGen Biotech, catalog number: CT10102 )
- Trans2K® Plus II DNA Marker (TransGen Biotech, catalog number: BM121-01 )
- Diethyl pyrocarbonate/DEPC (Sigma-Aldrich, catalog number: D5758 )
- TIANgel Midi Purification Kit (TransGen Biotech, catalog number: DP209 )
- UltraPureTM Agarose (Life Technologies, catalog number: 75510019 )
- DEPC-treated deionized water (see Recipes)
- 3 M sodium acetate (see Recipes)
- 70%, 100% ethanol (see Recipes)
- 50x TAE buffer (see Recipes)
Equipment
- MaxyClear Microtubes (1.5 ml) (Corning Incorporated, Axygen®, catalog number: MCT150C )
- NanoDropTM 1000 Spectrophotometer (Thermo Fisher Scientific, catalog number: ND-1000 )
- Thermomixer comfort (Eppendorf, catalog number: 5362.000.019 )
- Eppendorf Centrifuge (Eppendorf, catalog number: 5417R )
- PCR system (Eastwin Life Science, catalog number: EDC-810 )
- Other standard laboratory equipment
Procedure
- Primer design
- In general, primers should be highly specific for the target sequences, able to form stable duplexes with their target sequences, and free of secondary structure.
- Three primers around 20~25 nucleotides in length should be synthesized. The GC content of these primers is designed as similar as possible.
Note: We recommend the GC ratio to be between 40%~55%. e.g., GC ratios of primers listed in Figure 2A are in this range. However, this parameter is not fixed and it depends on your gene. - The first gene-specific primer cRT (Figure 1) should be localized in the known region and used for reverse transcription (RT).
Note: cRT is of reverse orientation of the gene. Its localization is recommended within 100~200 nt from the anticipated 5’ end of the RNA. e.g., 18c is within 204 nt from the anticipated 5’ end of mature 18S rRNA (Figure 2A). - The other two primers, cF1 and cR1, localized in 5’ and 3’ sides of the cRT primer, respectively (Figure 1), are for primary PCR amplification of the 5’ and 3’ extremities of the target transcript. cF1 and cR1 anneal to sequences located 3’ and 5’ (with respect to cDNA not RNA) of cRT, respectively. Their localizations are recommended within ~100 nt from the anticipated 5’ and 3’ end of the RNA, respectively.
Note: Primers cF2 or cR2, immediately down-stream of the cF1 or cR1 primer, respectively (Figure 1), are needed for further nested PCR to improve PCR specificity. Primers cF2 and cR2 for nested PCR could be designed in known region of mature RNA or predicted region only exist in precursor RNA. Compared to cF1/cR1, each of these three primer combinations (cF1/cR2, cF2/cR1 and cF2/cR2) could be used for nested PCR.
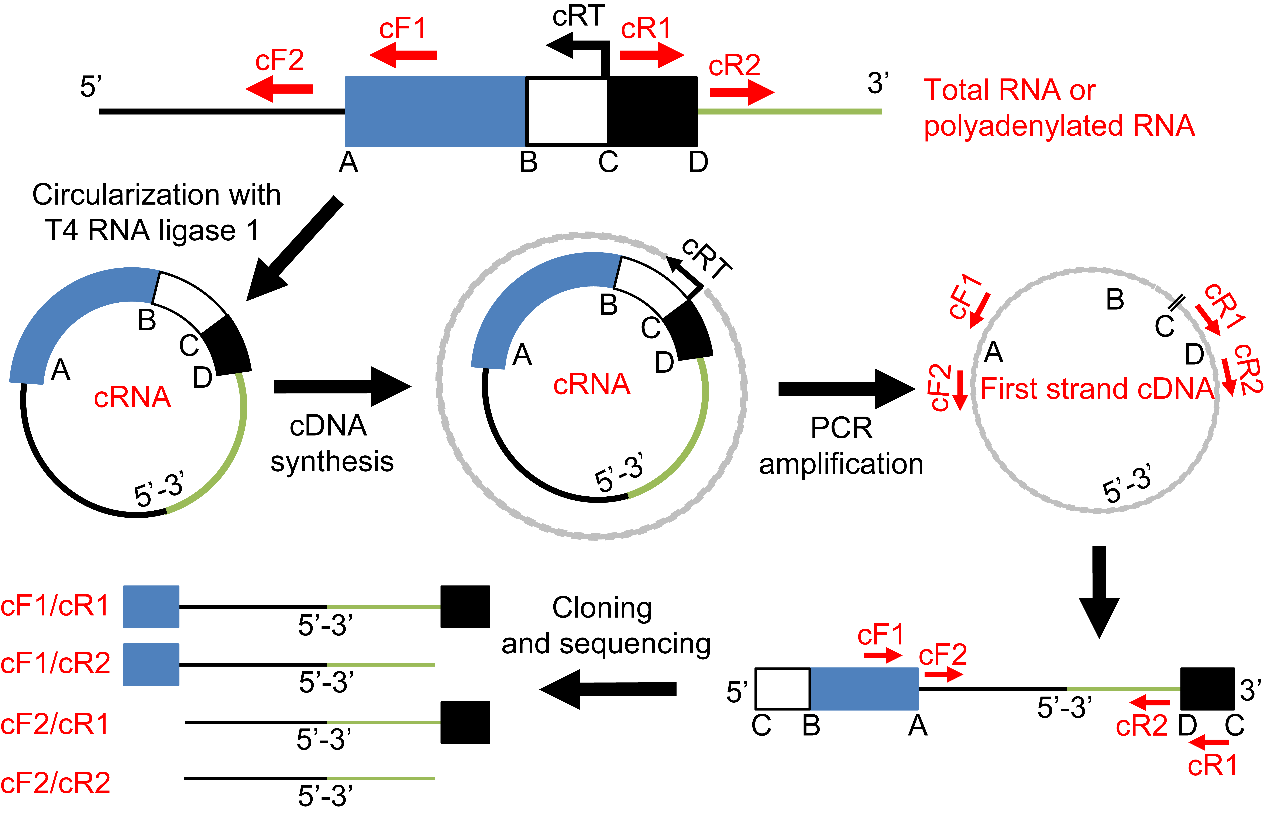
Figure 1. Diagrammatic representation of circular RT-PCR (cRT-PCR). Total RNA or polyadenylated RNA are circularized with T4 RNA ligase and reverse transcribed with gene-specific primer cRT. Then 5’ and 3’ extremities are amplified and sequenced. A and D mark the known 5’ and 3’ extremities of a specific RNA. Blue box from A to B and black box from C to D represent loci of PCR primers cF1 and cR1, respectively. White box between B and C indicates the locus of cRT primer. Primers cF2 and cR2 adjacent to the two primary primers cF1 and cR1, respectively, are designed for nested PCR. - In general, primers should be highly specific for the target sequences, able to form stable duplexes with their target sequences, and free of secondary structure.
- RNA extraction (Linear RNA)
- Total RNA is extracted from 0.1 g of 14-day-old fresh Arabidopsis seedlings with TRNzol Reagent according to standard manufacturer’s instruction, dissolved in DEPC-treated deionized water and quantified with NanoDropTM 1000 Spectrophotometer.
Note:At least 50 μg total RNA could be extracted from 0.1 g seedlings. Therefore in theory, 0.01 g seedlings may be sufficient for one RNA circularization reaction (10 μg RNA). In addition, the starting amount of plant tissue may vary depending on specific tissue type, since different tissues may need optimized protocols and reagents for better RNA extraction efficiency. - For RNAs with polyadenylated tails, enrichment with a Dynabeads® mRNA Purification Kit is recommended.
Note: In general, polyadenylations are often identified in 3’-end of most nucleus-encoded mRNAs in eukaryotes, and RNA molecules in prokaryotes and organelles (Slomovic et al., 2008). In addition, most of other non-coding RNAs are also found to be polyadenylated for processing or degradation, like rRNAs, snoRNA, snRNA and so on (Lykke-Andersen et al., 2009).
- Total RNA is extracted from 0.1 g of 14-day-old fresh Arabidopsis seedlings with TRNzol Reagent according to standard manufacturer’s instruction, dissolved in DEPC-treated deionized water and quantified with NanoDropTM 1000 Spectrophotometer.
- RNA ligation (Circular RNA, cRNA)
- 10 μg of total RNA or 0.5 μg of poly(A) RNA was used for each ligation reaction (50 μl). RNA is pre-incubated in Thermomixer comfort at 70 °C for 5 min, and then immediately chilled on ice for 2 min.
Note: RNA decapping step is recommended before RNA ligation when analyzing nucleus-encoded mRNAs (Slomovic et al., 2008), since RNAs with 5’-cap structure will inhibit the RNA ligation efficiency. Although this step was not applied in our previous work (Hang et al., 2014), we recommend performing this assay if necessary with these two kits according to their corresponding protocols. RNA 5´ Polyphosphatase from Epicentre
(http://www.epibio.com/docs/default-source/protocols/rna-5-polyphosphatase.pdf?sfvrsn=12) or Tobacco Acid Pyrophosphatase supported in FirstChoice® RLM-RACE Kit (https://tools.lifetechnologies.com/content/sfs/manuals/cms_056070.pdf). - Prepare the following reaction system:
RNA 10 μg total/0.5 μg poly(A)
T4 RNA Ligase 1 (10,000 U/ml) 3 μl (30 U)
10x T4 RNA Ligase 1 buffer5 μl
Adenosine-5'-Triphosphate (10 mM ATP) 5 μl (1 mM)
RNase inhibitor (40 U/μl) 1 μl (40 U)
DEPC-treated deionized waterto 50 μl
Note: The ligation reaction system may be optimized in the dosage of T4 RNA ligase 1 or ATP. Be aware that the appropriate concentration of ATP should be added if it is not supplied in the T4 RNA ligase buffer. The appropriate concentration of ATP should be in accordance with the product information of the commercial T4 RNA ligase 1. We used T4 RNA Ligase 1 from New England Biolabs (see in materials and reagents). In its properties and usage section, 1 mM ATP is suggested to add into the reaction system, since there is no ATP in the 10 x T4 RNA ligase reaction buffer (https://www.neb.com/products/m0204-t4-rna-ligase-1-ssrna-ligase - tabselect0). In addition, make reference to the companion protocol optimized for RNA circularization (https://www.neb.com/protocols/1/01/01/rna-circularization-using-t4-rna-ligase-1). In this protocol, 20 μl reaction system supplemented with 20-50 μM ATP and 10% PEG8000 is set up and then incubated at 25 °C for 1~2 h, or overnight at 16 °C for better yield. - Mix well and incubate in Thermomixer comfort at 37 °C for 1~2 h.
- Incubate at 65 °C for 15 min to inactivate the T4 RNA Ligase 1.
- Ethanol precipitate the RNA by adding 5 μl 3 M sodium acetate (pH 5.2), 2 μl of glycogen (20 mg/ml), and 171 μl of 100% ethanol. Keep the mixture on ice for 30 min (short time) or store at -20 °C overnight to precipitate the ligated RNA.
Note: Longer precipitation at -20 °C overnight is recommended for better RNA recovery. The concentration of glycogen is 20 mg/ml. Its working concentration here is around 0.7 μg/μl when 2 μl glycogen is added into 50 μl reaction product plus 5 μl 3M sodium acetate. It is notable that a final 0.05-1 μg/μl concentration of glycogen is suggested according to its product instruction. - Spin the sample at 12,000 rpm for 15 min at 4 °C.
Note:The force for a given rpm varies depending on the rotor. When we used Eppendorf Centrifuge 5417R, the speed 12,000 rpm equals to 15,294 x g. - Discard the supernatant and wash the pellet with 500 μl of 70% ethanol, then spin at 12,000 rpm for 15 min at 4 °C.
- Discard the supernatant and air-dry the pellet. Resuspend the RNA in DEPC-treated deionized water, 70 μl for total RNA or 7 μl for poly(A) RNA. RNA samples can be stored at -80 °C until used.
Note: We have not compared the half-life of the cRNA and normal RNA strored in -80 °C yet, although we predict that the cRNA is stable and could be kept for a long time in -80 °C. We recommend to perform the reverse transcription as soon as possible.
- 10 μg of total RNA or 0.5 μg of poly(A) RNA was used for each ligation reaction (50 μl). RNA is pre-incubated in Thermomixer comfort at 70 °C for 5 min, and then immediately chilled on ice for 2 min.
- Reverse transcription with gene-specific primer (Circular cDNA, ccDNA)
- Prepare the following reaction system and pre-incubate the mixture at 70 °C for 5 min, then chill it on ice for 2 min. 1 μg of total cRNA or 0.5 μg of poly(A) cRNA is used for each reverse transcription.
cRNA 7 μl
RT-primer (cRT in Figure 1)2 pmol
DEPC-treated deionized waterto 9 μl - Spin the mixture down slightly and add the following reaction mixture to each sample.
2x TS Reaction Mix10 μl
TransScript® II RT/RI Enzyme Mix 1 μl
Mix well and incubate at 50 °C for 1 h.
Note:The 2x TS Reaction Mix and TransScript® II RT/RI Enzyme Mix are components supported in TransScript® II First-Strand cDNA Synthesis SuperMix. They have no catalog numbers and therefore not listed in the materials and reagents section. - Incubate at 85 °C for 5 min to inactivate the TransScript® II RT enzyme.
- Add 180 μl of DEPC-treated deionized water to dilute the product to the final volume of 200 μl.
- Prepare the following reaction system and pre-incubate the mixture at 70 °C for 5 min, then chill it on ice for 2 min. 1 μg of total cRNA or 0.5 μg of poly(A) cRNA is used for each reverse transcription.
- PCR amplification
- Prepare the general reaction mixture for PCR (50 μl in total):
cDNA5 μl
ExTaq 10x buffer5 μl
dNTPs mix (2.5 mM each dNTP)4 μl
Forward primer (cF1, 10 μM)1 μl (10 pmol)
Reverse primer (cR1, 10 μM)1 μl (10 pmol)
TaKaRa ExTaq® DNA Polymerase (5 U/μl)0.12 μl (0.6 U)
Deionized water 33.88 μl - Run the PCR amplification according to the program described below:
a.3 min at 94 °C;
b.30 sec at 94 °C;
c.30 sec at 55 °C;
d.1 min at 72 °C; Repeat steps b-d for 30 cycles
e.10 min at 72 °C;
f. Hold at 20 °C.
Note: Annealing temperature and elongation time could be optimized case by case. We recommend to try the routine annealing temperature between 55~60 °C first. As we know, higher annealing temperature may increase the product specificity. Therefore, we could compare the PCR products in 55 °C, 58 °C and 60 °C to choose the best one. For the elongation time, we prefer to choose 1 min for the first try to have a look at the PCR product length (we suppose it isn’t longer than 1,000 bp). Then, we will adjust the elongation time accordingly. In summary, the elongation time depends on the potential length of PCR products and elongation rate of DNA polymerase. - Load 8 μl of the PCR products on 1% agarose gel (with Ethidium Bromide, EtBr) and run until the bromophenol blue dye is 2/3rds of the way down the gel. Image the gel and determine whether this batch of circular RT-PCR is successful (sharp and strong bands) and which candidate band should be cut for further analysis.
Note: If it does not work well, PCR system should be optimized in annealing temperature, amplification cycles, PCR enzyme, and so on. In addition, on the basis of the primary PCR with cF1/cR1, a further nested PCR with cF1/cR2, cF2/cR1 or cF2/cR2 could be helpful to improve the product specificity as described in Figure 1. For the nested PCR, the PCR mix components and PCR program are the same as the first round PCR, except for the primer combination and DNA template. - Load all the remaining PCR products on a new 1% agarose gel. Cut the candidate bands and purify the DNA from the gel with TIANgel Midi Purification Kit.
- Prepare the general reaction mixture for PCR (50 μl in total):
- DNA sequencing and analysis
- Purified DNA products are cloned into the pEasy-T vector.
- Make a master reaction mix for colony PCR, except the DNA template (20 μl in total):
ExTaq 10x buffer2 μl
dNTPs mix (2.5 mM each dNTP)1 μl
M13F primer (10 μM)0.5 μl (5 pmol)
M13R primer (10 μM)0.5 μl (5 pmol)
TaKaRa ExTaq® DNA Polymerase (5 U/μl)0.12 μl (0.6 U)
Deionized water 15.88 μl
PCR reaction mix is prepared first and aliquoted into 0.2 ml PCR tubes. - Positive clones are chosen by white-blue colony selection. Take a 20 μl pipet tip and touch a positive colony very lightly and dip the tip a couple of times into the 20 μl of PCR mix.
- Run the PCR amplification according to the program described in E.2. The elongation time depends on the length of PCR product. For TaKaRa ExTaq® DNA Polymerase, the elongation time is 1 min for 1 kbp PCR products.
- Analyze the PCR products using a 1% agarose gel with EtBr staining. Excise gel slices containing sharp target bands for DNA sequencing with M13F sequencing primer. We recommend to sequence at least 15 samples for a reliable result.
Note: DNA sequences with both M13F and M13R primers are necessary to determine the 5’ and 3’ extremities of the target RNAs, especially for highly polyadenylated RNAs at the 3’ tail. - DNA sequencing data are analyzed with Basic Local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information (NCBI). Choose the organism: Arabidopsis thaliana. Search database: Reference genomic sequences (refseq_genomic) using Megablast (Optimized for highly similar sequences).
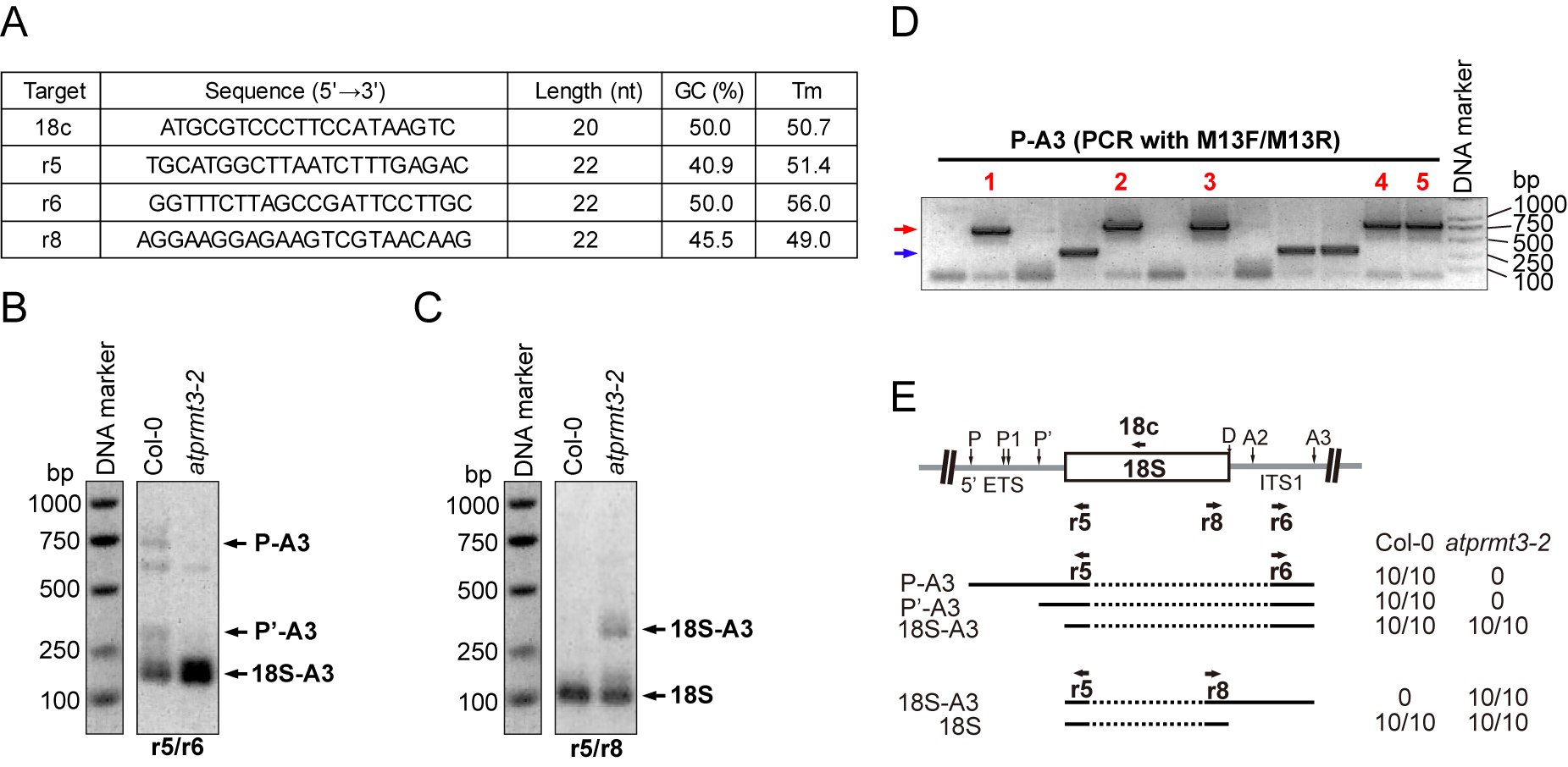
Figure 2. Circular RT-PCR to identify the pre-18S rRNAs in atprmt3-2. Total RNA from 14-day-old seedlings of both Col-0 and atprmt3-2 mutants are used for cRT-PCR assay. (A) Oligonucleotides used in this assay. 18c is the cRT primer for reverse transcription of pre-18S rRNAs. Their lengths, GC content and Tm value are listed. (B-C) Agarose gel analysis of PCR products amplified with primers r5/r6 (B) and r5/r8 (C) after reverse transcription with 18c. Reaction products were analyzed by gel electrophoresis in a 1.5% agarose gels and stained with 0.5 μg/ml ethidium bromide. DNA markers are indicated on the left. Excise candidate bands marked on the right and extract DNAs to clone into the pEasy-T vector. (D) Perform colony PCR with M13F and M13R primers for 30 cycles. Excise target bands with the right molecular weight (red arrow) for DNA sequencing with M13F sequencing primer. Avoid the PCR products with abnormal molecular weight (blue arrow), which are supposed to be contaminations in the gel-excising step. (E) The diagram illustrates the primers’ localizations, the amplified PCR fragments and the DNA sequencing results. For each fragment, the number of clones obtained from wild-type and atprmt3-2 samples is indicated on the right. 5’ ETS, external transcribed sequence. ITS1, internal transcribed sequence. P, P1 and P’ are known cleavage sites in 5’ ETS. D, A2 and A3 are known cleavage sites in ITS1. - Purified DNA products are cloned into the pEasy-T vector.
Recipes
- DEPC-treated deionized water
Add 1 ml DEPC to 1 L deionized water, mix and put at room temperature overnight
Autoclave at 120 °C for 30 min - 3 M sodium acetate (pH 5.2)
Dissolve 40.8 g sodium acetate in 40 ml DEPC-treated deionized water, adjust to pH 5.2 with acetic acid.
Add deionized water to the final volume of 100 ml
Autoclave at 120 °C for 15 min - 70%, 100% ethanol
Diluted with DEPC-treated deionized water - 50x TAE buffer
Dissolve 242 g Tris base and 37.2 g Na2EDTA.2H2O in 800 ml deionized water, add 57.1 ml acetic acid and adjust to pH 8.0 with NaOH.
Add deionized water to the final volume of 1,000 ml
Acknowledgments
This protocol was adapted from the previously published studies (Barkan, 2011; Slomovic et al., 2008) and Dr. Alice Barkan’s lab homepage (http://pml.uoregon.edu/protocols.html), and it was performed in (Hang et al., 2014). This work was supported by National Natural Science Foundation of China Grants 31330020 and 31210103901 (to X.C.), 31370770 and 31171184 (to C.L.), and 31200900 (to X.D.), and State Key Laboratory of Plant Genomics Grant SKLPG2011B0101.
References
- Barkan, A. (2011). Studying the structure and processing of chloroplast transcripts. Methods Mol Biol 774: 183-197.
- Hang, R., Liu, C., Ahmad, A., Zhang, Y., Lu, F. and Cao, X. (2014). Arabidopsis protein arginine methyltransferase 3 is required for ribosome biogenesis by affecting precursor ribosomal RNA processing. Proc Natl Acad Sci U S A 111(45): 16190-16195.
- Lykke-Andersen, S., Brodersen, D. E. and Jensen, T. H. (2009). Origins and activities of the eukaryotic exosome. J Cell Sci 122(Pt 10): 1487-1494.
- Slomovic, S., Portnoy, V. and Schuster, G. (2008). Detection and characterization of polyadenylated RNA in Eukarya, Bacteria, Archaea, and organelles. Methods Enzymol 447: 501-520.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hang, R., Deng, X., Liu, C., Mo, B. and Cao, X. (2015). Circular RT-PCR Assay Using Arabidopsis Samples. Bio-protocol 5(14): e1533. DOI: 10.21769/BioProtoc.1533.
Category
Plant Science > Plant molecular biology > RNA > Transcription
Molecular Biology > RNA > qRT-PCR
Molecular Biology > DNA > PCR
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










