- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Identification of Proteins Interacting with Genomic Regions of Interest in vivo Using Engineered DNA-binding Molecule-mediated Chromatin Immunoprecipitation (enChIP)
Published: Vol 4, Iss 10, May 20, 2014 DOI: 10.21769/BioProtoc.1124 Views: 15459
Reviewed by: Sabine Le SauxAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

RNA Chromatin Immunoprecipitation (RNA-ChIP) in Caenorhabditis elegans
Germano Cecere and Alla Grishok
Dec 20, 2014 15654 Views

Chromatin Immunoprecipitation (ChIP) Assay for Detecting Direct and Indirect Protein – DNA Interactions in Magnaporthe oryzae
Gang Li [...] Richard A. Wilson
Nov 5, 2015 15982 Views
Abstract
Elucidation of molecular mechanisms of genome functions requires identification of molecules interacting with genomic regions of interest in vivo. To this end, it is useful to isolate the target regions retaining molecular interactions. We established locus-specific chromatin immunoprecipitation (ChIP) technologies consisting of insertional ChIP (iChIP) and engineered DNA-binding molecule-mediated ChIP (enChIP) for isolation of target genomic regions (Hoshino and Fujii, 2009; Fujita and Fujii, 2011; Fujita and Fujii, 2012; Fujita and Fujii, 2013a; Fujita and Fujii, 2013b; Fujita et al., 2013). Identification and characterization of molecules interacting with the isolated genomic regions facilitates understanding of molecular mechanisms of functions of the target genome regions.
Here, we describe enChIP, in which engineered DNA-binding molecules, such as zinc-finger proteins, transcription activator-like (TAL) proteins, and a catalytically inactive Cas9 (dCas9) plus small guide RNA (gRNA), are utilized for affinity purification of target genomic regions. The scheme of enChIP is as follows:
1. A zinc-finger protein, TAL or dCas9 plus gRNA is generated to recognize DNA sequence in a genomic region of interest.
2. The engineered DNA-binding molecule is fused with a tag(s) and the nuclear localization signal (NLS), and expressed in the cell to be analyzed.
3. The resultant cell is crosslinked, if necessary, and lysed, and DNA is fragmented.
4. The complexes including the engineered DNA-binding molecule are subjected to affinity purification such as mmunoprecipitation. The isolated complexes retain molecules interacting with the genomic region of interest.
5. Reverse crosslinking and subsequent purification of DNA, RNA, or proteins allow identification and characterization of these molecules.
In this protocol, we describe enChIP with a TAL protein to isolate a genomic region of interest and analyze the interacting proteins by mass spectrometry (Fujita et al., 2013).
Materials and Reagents
- Target cells
- 37% formaldehyde (Nacalai Tesque, catalog number: 16223-55 )
- Glycine (Sigma-Aldrich, catalog number: G7126 )
- NaCl (Sigma-Aldrich, catalog number: S9625 )
- Agarose S (Wako Pure Chemical Industries, catalog number: 318-01195 )
- 1 M Tris (pH 6.8) (AppliChem GmbH, catalog number: A4987 )
- 1 M Tris (pH 7.5) (AppliChem GmbH, catalog number: A4263 )
- 1 M Tris (pH 8.0) (AppliChem GmbH, catalog number: A4577 )
- 0.5 M EDTA (pH 8.0) (Nacalai Tesque, catalog number: 14362-95 )
- 0.1 M EGTA (Nacalai Tesque, catalog number: 08947-35 )
- IGEPAL CA-630 (Sigma-Aldrich, catalog number: I8896 )
- Triton X-100 (Nacalai Tesque, catalog number: 25987-85 )
- 10% SDS solution (Nacalai Tesque, catalog number: 30562-04 )
- 8 M LiCl solution (Nacalai Tesque, catalog number: 20077-84 )
- 2-mercaptoethanol (Nacalai Tesque, catalog number: 21438-82 )
- Sucrose (Nacalai Tesque, catalog number: 30404-45 )
- Bromophenol blue (Nacalai Tesque, catalog number: 05808-61 )
- Tween-20 (Sigma-Aldrich, catalog number: P5927 )
- Deoxycholic acid sodium salt monohydrate (Nacalai Tesque, catalog number: 10712-12 )
- 30% lauroylsarcosine (Nacalai Tesque, catalog number: 20135-14 )
- Dynabeads-protein G (Life Technologies, catalog number: DB10004 )
- Anti-FLAG M2 antibody (Sigma-Aldrich, catalog number: F1804 )
- Normal mouse IgG (Santa Cruz, catalog number: sc-2025 )
- 10x phosphate buffered saline (PBS) (pH7.4) (Nacalai Tesque, catalog number: 27575-31 )
- PBS (10x dilution of 10x PBS with distilled water)
- BSA fraction V (7.5%) (Life Technologies, catalog number: 15260 )
- Complete, mini, EDTA-free protease inhibitor (Roche Diagnostics, catalog number: 4693159 )
- 10 mg/ml RNase A (Sigma-Aldrich, catalog number: R6513 )
- 20 mg/ml Proteinase K (Roche Diagnostics, catalog number: 3115828 )
- 3x FLAG peptide (Sigma-Aldrich, catalog number: F4799 )
- 2-propanol (Nacalai Tesque, catalog number: 29113-95 )
- 3 M sodium acetate buffer solution (pH 5.2) (Nacalai Tesque, catalog number: 06893-24 )
- 20 mg/ml glycogen (Roche Diagnostics, catalog number: 901393 )
- 70% ethanol
- 4-20% mini-PROTEAN TGX gel (10 well, 50 µl) (Bio-Rad Laboratories, catalog number: 456-1094 )
- 10x running buffer solution for SDS-PAGE (Tris-Glycine) (Nacalai Tesque, catalog number: 30329-74 )
- Coomassie brilliant blue R-250 staining solution (Bio-Rad Laboratories, catalog number: 161-0436 )
- In-Gel Tryptic Digestion Kit (Thermo Fisher Scientific, catalog number: 89871 )
- 1.25 M glycine solution (200 ml) (see Recipes)
- 10% sodium deoxycholate (10 ml) (see Recipes)
- 10% Tween-20 (10 ml) (see Recipes)
- Cell lysis buffer (CLB) (40 ml) (see Recipes)
- Nuclear lysis buffer (NLB) (40 ml) (see Recipes)
- Modified lysis buffer 3 (MLB3) (10 ml) (see Recipes)
- PBS-T (10 ml) (see Recipes)
- PBS-T-BSA (10 ml) (see Recipes)
- 5% Triton X-100 (in MLB3) (5 ml) (see Recipes)
- Low salt buffer (LSB) (10 ml) (see Recipes)
- High salt buffer (HSB) (10 ml) (see Recipes)
- LiCl buffer (20 ml) (see Recipes)
- TBS-IGEPAL CA-630 (10 ml) (see Recipes)
- Elution buffer (500 µl) (see Recipes)
- 2x sample buffer (50 ml) (see Recipes)
Equipment
- Magnetic stand (magical trapper) (TOYOBO, catalog number: MGS-101 )
- Centrifuge
- 1.5 ml centrifuge tube (Sarstedt AG, catalog number: 72.690.001 )
- 2 ml centrifuge tube (Eppendorf, catalog number: 0030.120.094 )
- 50 ml centrifuge tube (BD Biosciences, catalog number: 352070 )
- Vortex mixer
- Rotator
- Vaccum pump connected to a side-arm flask
- Sonicator (ultrasonic disruptor UD-201 ) (TOMY SEIKO, catalog number: UD-201)
- Shaker
- Mass spectrometry facility: a nanoLC-MS/MS system, composed of LTQ Orbitrap Velos (Thermo Fisher Scientific) coupled with nanoLC (Advance CaptiveSpray SOURCE, Michrom BioResources) and HTC-PAL autosampler (CTC Analytics)
Procedure
- Preparation of cells
- Design the DNA-binding modules of a TAL protein recognizing a specific sequence (approximately 20 bases) in your target genomic region.
- Express the TAL protein fused with 3x FLAG tags and the NLS in the cell to be analyzed. An example of the tagged TAL proteins for enChIP is shown in supplemental Figure 1 in the reference (Fujita et al., 2013).
- Design the DNA-binding modules of a TAL protein recognizing a specific sequence (approximately 20 bases) in your target genomic region.
- Formaldehyde crosslinking of cells
- Culture target cells. Use 2 x 107 cells x 2 (total 4 x 107 cells) (e.g. Ba/F3) for chromatin preparation. Collect cells suspended in culture medium (2 x 107 cells/30 ml) in a 50 ml centrifuge tube (if necessary, increase cell number). Prepare 2 tubes (total 4 x 107 cells).
- Add 810 µl of 37% formaldehyde to 1% final concentration into 30 ml of the cell suspension. Formaldehyde solution is harmful, so handle it wearing gloves in a chemical hood. Incubate at 37 °C for 5-10 min (usually 5 min). Crosslinking makes color of cell suspension slightly yellowish.
- Stop crosslinking by adding 3.1 ml of 1.25 M glycine solution to 127 mM final concentration. Incubate at room temperature for 10 min.
- Collect cells by centrifugation (300 x g, 4 °C, 5 min). Discard carefully the supernatant including formaldehyde by decantation, and store it in a waste bottle.
- Resuspend the fixed cells (pellet) in 30 ml of PBS by vortexing. Collect cells by centrifugation (300 x g, 4 °C, 5 min). Discard carefully the supernatant including formaldehyde by decantation, and store it in a waste bottle.
- Repeat the step B5 (total twice). The formaldehyde wastes should be handled according to the chemical safety guideline of your laboratory.
- Proceed to the step C1, or the centrifuge tube with the fixed cells (pellet) can be frozen and stored at -80 °C.
- Culture target cells. Use 2 x 107 cells x 2 (total 4 x 107 cells) (e.g. Ba/F3) for chromatin preparation. Collect cells suspended in culture medium (2 x 107 cells/30 ml) in a 50 ml centrifuge tube (if necessary, increase cell number). Prepare 2 tubes (total 4 x 107 cells).
- Preparation of chromatin (per 2 x 107 cells)
- Resuspend the fixed cells in 10 ml of CLB. If the fixed cells are frozen in the step B6, it should be thawed before addition of CLB. Incubate on ice for 10 min.
- Centrifuge at 930 x g at 4 °C for 8 min. Discard carefully the supernatant by vacuum suction.
- Resuspend the pellet in 10 ml of NLB. Incubate on ice for 10 min. Vortex at maximum setting for 2-3 sec every 2-3 min.
- Centrifuge at 930 x g at 4 °C for 8 min. Discard carefully the supernatant by vacuum suction.
- Resuspend the pellet in 10 ml of PBS. Centrifuge at 930 x g at 4 °C for 10 min. Discard carefully the supernatant by vacuum suction. The pellet is used as the chromatin fraction.
- Proceed to the step D1 or the centrifuge tube with the chromatin fraction can be stored at -80 °C after immediate freezing in liquid nitrogen.
- Resuspend the fixed cells in 10 ml of CLB. If the fixed cells are frozen in the step B6, it should be thawed before addition of CLB. Incubate on ice for 10 min.
- Sonication of chromatin (per 2 x 107 cells)
- Resuspend the collected chromatin fraction in 800 µl of MLB3. Transfer the suspension into a 1.5 ml microtube.
- Sonicate the chromatin by using ultrasonic disruptor UD-201. Conditions are as follows:
Output: 3
Duty: 100% (continuous)
Time: Free
10-18 cycles of sonication for 10 sec and cooling on ice for 20 sec
2 min on ice after 5-6 cycles to avoid excessive heating
Keep the position of the tip of the sonication bar approximately 0.5 cm away from the tube bottom using a clamp or hands to avoid foaming. - Centrifuge at 16,000 x g at 4 °C for 10 min. Transfer the supernatant (800 µl) into a 1.5 ml tube. The 1.5 ml tube with the supernatant can be stored at -80 °C after immediate freezing in liquid nitrogen.
- Resuspend the collected chromatin fraction in 800 µl of MLB3. Transfer the suspension into a 1.5 ml microtube.
- Evaluation of fragmentation of chromatin
- Resuspend 10 µl of the fragmented chromatin in 85 µl of distilled water.
- Add 4 µl of 5 M NaCl. Incubate at 65 °C overnight.
- Add 1 µl of 10 mg/ml RNase A. Incubate at 37 °C for 45 min.
- Add 2 µl of 0.5 M EDTA (pH 8.0), 4 µl of 1 M Tris (pH 6.8), and 1 µl of 20 mg/ml Proteinase K. Incubate at 45 °C for 1.5 h.
- Prepare 1% agarose gel without staining dye.
- Use 10 µl of the sample prepared in the step E4 for electrophoresis in 1% agarose gel without staining dye. Run the gel at 100 V for 30 min. 1 kbp DNA ladder marker should be loaded to estimate DNA fragmentation.
- Stain the gel with staining dye (e.g. ethidium bromide) for 0.5-1 h (Figure 1A). An example of DNA fragmentation is shown in Figure 1B. The acceptable range of DNA fragmentation is dependent on the length of your target genomic region. Usually, the range of DNA fragmentation should be generally 4-0.2 kbp (The average length of DNA fragments should be 0.5-2 kbp). If DNA is poorly or excessively fragmented, adjust output or cycles of sonication in the step D2.
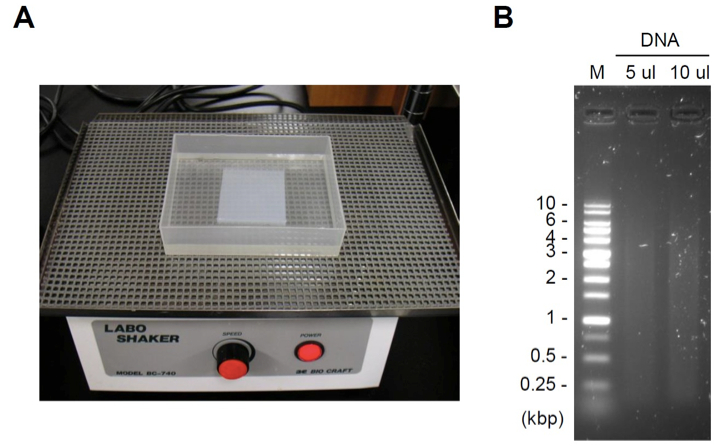
Figure 1. Gel staining and an example of DNA fragmentation. A. Gel staining with staining dye using a shaker (speed: one turn/1-2 sec). B. An example of DNA fragmentation. M: DNA ladder marker.
- Resuspend 10 µl of the fragmented chromatin in 85 µl of distilled water.
- Preparation of Dynabeads conjugated with antibody (anti-FLAG antibody or normal mouse IgG)
- Prepare two 2 ml tubes; one is for anti-FLAG antibody and the other is for normal mouse IgG. Add 300 µl of Dynabeads-Protein G in each tube (300 µl x 2 tubes). In this protocol, we describe preparation of one sample for analysis by mass spectrometry. In this regard, normal mouse IgG is used for pre-clear in the step G3 and anti-FLAG antibody is used for affinity purification in the step G5. If you analyze another sample as a negative control (e.g. affinity purification with normal mouse IgG instead of anti-FLAG antibody), you may prepare chromatin and Dynabeads for it separately.
- Put the tubes on a magnet stand and wait for 3 min. Discard the supernatant by pipetting. Handling of a magnet stand is shown in Figure 2.
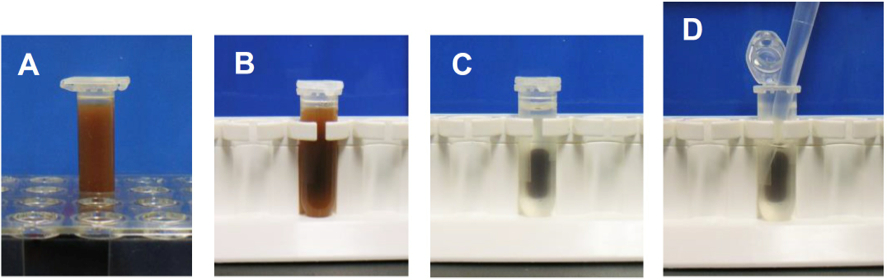
Figure 2. Handling of a magnet stand in wash steps. A. Suspension of Dynabeads in a 2 ml tube. B. Setting of the tube on a magnet stand. C. 3 min after the setting. D. Removal of the supernatant by pipetting. - Resuspend the pellet in 1 ml of PBS-T. Put the tubes on a magnet stand and wait for 2 min. Discard the supernatant by pipetting.
- Repeat the step F3 (total twice).
- Resuspend the pellet in 1.5 ml of PBS-T-BSA.
- Add 30 µg of antibody. One tube is for 30 µg of anti-FLAG antibody, and the other is for 30 µg of normal mouse IgG. Rotate at 4 °C overnight.
- Centrifuge briefly (400 x g for 2-3 sec). Put the tubes on a magnet stand and wait for 3 min. Discard the supernatant by pipetting.
- Resuspend the pellet in 1.5 ml of PBS-T. Invert several times and centrifuge briefly (400 x g for 2-3 sec). Put the tubes on a magnet stand and wait for 3 min. Discard the supernatant by pipetting.
- Repeat the step F8, twice (total three times). The Dynabeads are ready for the next step.
- Prepare two 2 ml tubes; one is for anti-FLAG antibody and the other is for normal mouse IgG. Add 300 µl of Dynabeads-Protein G in each tube (300 µl x 2 tubes). In this protocol, we describe preparation of one sample for analysis by mass spectrometry. In this regard, normal mouse IgG is used for pre-clear in the step G3 and anti-FLAG antibody is used for affinity purification in the step G5. If you analyze another sample as a negative control (e.g. affinity purification with normal mouse IgG instead of anti-FLAG antibody), you may prepare chromatin and Dynabeads for it separately.
- Chromatin immunoprecipitation
- Transfer 1.6 ml of the fragmented chromatin, which corresponds to chromatin extracted from 4 x 107 cells, into a 2 ml tube.
- Add 400 µl of 5% Triton X-100 (in MLB3) to 1% final concentration.
- Add chromatin solution prepared in the step G2 (2 ml) in the tube, in which the Dynabeads conjugated with normal mouse IgG were prepared. Rotate at 4 °C for 1 h.
- Put the tube on a magnet stand and wait for 3 min.
- Transfer the supernatant into the tube, in which the Dynabeads conjugated with anti-FLAG antibody were prepared. Rotate at 4 °C overnight.
- Put the tube on a magnet stand and wait for 3 min. Discard the supernatant by pipetting.
- Wash 1: Add 1.8 ml of LSB. Rotate at 4 °C for 5 min. Put the tube on a magnet stand and wait for 3 min. Discard the supernatant by pipetting.
- Repeat wash 1 (total twice).
- Wash 2: Wash with HSB as described in the step G7. Repeat this wash (total twice).
- Wash 3: Wash with LiCl buffer as described in the step G7. Repeat this wash (total twice).
- Add 1.8 ml of TBS-IGEPAL CA-630. Rotate at 4 °C for 5 min. Put the tube on a magnet stand and wait for 3 min. Discard the supernatant by pipetting.
- Elution: Resuspend the pellet in 200 µl of elution buffer. Incubate at 37 °C for 20 min. Put the tube on a magnet stand and wait for 3 min.
- Add the supernatant (200 µl) in the 1.5 ml tube with 0.5 ml of 2-propanol, 25 µl of 3 M Sodium acetate buffer solution (pH 5.2), and 5 µl of 20 mg/ml glycogen. Precipitate proteins at -20 °C overnight.
- Centrifuge at 16,000 x g at 4 °C for 30 min. Discard the supernatant.
- Rinse with 1 ml of 70% ethanol. Centrifuge at 16,000 x g at 4 °C for 10 min. Discard the supernatant completely by pipetting.
- Add 40 µl of 2x sample buffer. Vortex at maximum setting for 5 min to completely dissolve the precipitate. Incubate at 100 °C for 30 min (protein denaturing and reverse-crosslinking).
- Transfer 1.6 ml of the fragmented chromatin, which corresponds to chromatin extracted from 4 x 107 cells, into a 2 ml tube.
- SDS-PAGE, staining, mass analysis
- SDS-PAGE. Run the eluate on polyacrylamide gel until the dye reaches 1 cm from the well.
- Coomassie brilliant blue (CBB) staining or silver staining. An example of gel images is shown in Figure 3.
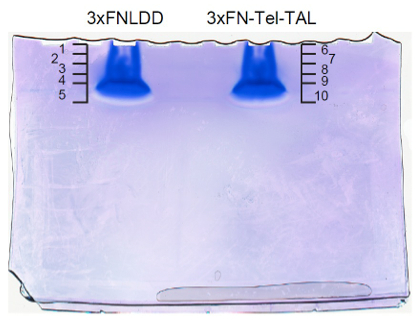
Figure 3. An example of gel images after CBB staining. The original image is shown in Supplemental Figure 5 in the reference (Fujita et al., 2013). - Cut the gel into 5 pieces x 2 mm.
- In gel digestion and mass analysis. You can use In-Gel Tryptic Digestion Kit. Our current system of mass analysis is a nanoLC-MS/MS system, composed of LTQ Orbitrap Velos coupled with nanoLC and HTC-PAL autosampler.
- SDS-PAGE. Run the eluate on polyacrylamide gel until the dye reaches 1 cm from the well.
Recipes
- 1.25 M glycine solution (200 ml)
Glycine (MW: 75.07) 18.8 g H2O to 200 ml - 10% sodium deoxycholate (10 ml)
Deoxycholic acid sodium salt monohydrate 1 g H2O to 10 ml - Cell lysis buffer (CLB) (40 ml)
10 mM Tris (pH 8.0), 1 mM EDTA, 0.5% IGEPAL CA-630, 1x protease inhibitors1 M Tris (pH 8.0) 400 µl 0.5 M EDTA 80 µl IGEPAL CA-630 200 µl Complete, mini, EDTA-free 4 tablets Double distilled water (DDW) 39.32 ml - Nuclear lysis buffer (NLB) (40 ml)
10 mM Tris (pH 8.0), 1 mM EDTA, 0.5 M NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.5% lauroylsarcosine, 1x protease inhibitors1 M Tris (pH 8.0) 400 µl 0.5 M EDTA 80 µl 5 M NaCl 4 ml Triton X-100 400 µl 10% sodium deoxycholate 2 ml 30% lauroylsarcosine 666 µl Complete, mini, EDTA-free 4 tablets DDW 32.46 ml - Modified lysis buffer 3 (MLB3) (10 ml)
10 mM Tris (pH 8.0), 1 mM EDTA, 0.5 mM EGTA, 150 mM NaCl, 0.1% sodium deoxycholate, 0.1% SDS, 1x protease inhibitors1 M Tris (pH 8.0) 100 µl 0.5 M EDTA 20 µl 0.1 M EGTA 50 µl 5 M NaCl 300 µl 10% sodium deoxycholate 100 µl 10% SDS 100 µl Complete, mini, EDTA-free 1 tablet DDW 9.33 ml - 10% Tween-20 (10 ml)
Tween-20 1 ml DDW 9 ml - PBS-T (10 ml)
PBS (pH 7.4), 0.01% Tween-20PBS 10 ml 10% Tween-20 10 µl - PBS-T-BSA (10 ml)
PBS (pH 7.4), 0.01% Tween-20, 0.1% BSAPBS 10 ml 10% Tween-20 10 µl 7.5% BSA fraction V 133 µl - 5% Triton X-100 (in MLB3) (5 ml)
MLB3 4.75 ml Triton X-100 250 µl - Low salt buffer (LSB) (10 ml)
20 mM Tris (pH 8.0), 2 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.1% SDS1 M Tris (pH 8.0) 200 µl 0.5 M EDTA 40 µl 5 M NaCl 300 µl Triton X-100 100 µl 10% SDS 100 µl DDW 9.26 ml - High salt buffer (HSB) (10 ml)
20 mM Tris (pH 8.0), 2 mM EDTA, 500 mM NaCl, 1% Triton X-100, 0.1% SDS1 M Tris (pH 8.0) 200 µl 0.5 M EDTA 40 µl 5 M NaCl 1 ml Triton X-100 100 µl 10% SDS 100 µl DDW 8.56 ml - LiCl buffer (20 ml)
10 mM Tris (pH 8.0), 1 mM EDTA, 0.25 M LiCl, 0.5% IGEPAL CA-630, 0.5% sodium deoxycholate1 M Tris (pH 8.0) 200 µl 0.5 M EDTA 40 µl 8 M LiCl 625 µl IGEPAL CA-630 100 µl 10% sodium deoxycholate 1 ml DDW 18.035 ml - TBS-IGEPAL CA-630 (10 ml)
50 mM Tris (pH 7.5), 150 mM NaCl, 0.1% IGEPAL CA-6301 M Tris (pH 7.5) 500 µl 5 M NaCl 300 µl IGEPAL CA-630 10 µl DDW 9.19 ml - Elution buffer (500 µl)
50 mM Tris (pH 7.5), 150 mM NaCl, 0.1% IGEPAL CA-630, 500 µg/ml 3x FLAG peptide5 mg/ml 3x FLAG peptide in TBS 50 µl TBS-IGEPAL CA-630 450 µl - 2x sample buffer (50 ml)
125 mM Tris (pH 6.8), 10% 2-mercaptoethanol, 4% SDS, 10% sucrose, 0.004% bromophenol blue1 M Tris-HCl (pH 6.8) 6.25 ml 2-mercaptoethanol 5 ml SDS 2 g Sucrose 5 g Bromophenol blue 2 mg DDW to 50 ml
Acknowledgments
This protocol was adapted from the previously published paper (Fujita et al., 2013). We thank F. Kitaura for technical assistance. This work was supported by Research Institute for Microbial Diseases (T.F. and H.F.), Program for Combined Research Fields from Immunology Frontier Research Center, Osaka University (H.F.), the Takeda Science Foundation (H.F. and T.F.), the Asahi Glass Foundation (H.F.), A-STEP from Japan Science and Technology Agency (H.F.), Grant-in-Aid for Young Scientists (B) (#22710185) (T.F.), Grant-in-Aid for Scientific Research on Innovative Areas ‘‘Cell Fate’’ (#23118516) (T.F.), ‘‘The Genofield’’ (#23114707) (H.F.), ‘‘Transcription Cycle’’ (25118512) (H.F.).
References
- Fujita, T. and Fujii, H. (2011). Direct identification of insulator components by insertional chromatin immunoprecipitation. PLoS One 6(10): e26109.
- Fujita, T. and Fujii, H. (2012). Efficient isolation of specific genomic regions by insertional chromatin immunoprecipitation (iChIP) with a second-generation tagged LexA DNA-binding domain. Advances Biosci Biotechnol 3(5): 626-629.
- Fujita, T. and Fujii, H. (2013a). Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem Biophys Res Commun 439(1): 132-136.
- Fujita, T. and Fujii, H. (2013b). Locus-specific biochemical epigenetics/chromatin biochemistry by insertional chromatin immunoprecipitation. ISRN Biochem 2013, Article ID 913273.
- Fujita, T., Asano, Y., Ohtsuka, J., Takada, Y., Saito, K., Ohki, R. and Fujii, H. (2013). Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Sci Rep 3: 3171.
- Hoshino, A. and Fujii, H. (2009). Insertional chromatin immunoprecipitation: a method for isolating specific genomic regions. J Biosci Bioeng 108(5): 446-449.
Article Information
Copyright
© 2014 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Fujita, T. and Fujii, H. (2014). Identification of Proteins Interacting with Genomic Regions of Interest in vivo Using Engineered DNA-binding Molecule-mediated Chromatin Immunoprecipitation (enChIP). Bio-protocol 4(10): e1124. DOI: 10.21769/BioProtoc.1124.
Category
Molecular Biology > DNA > DNA-protein interaction
Biochemistry > Protein > Immunodetection > ChIP
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











