Advanced Search
Cortical excitatory neuron differentiation
Last updated date: Mar 29, 2021 Views: 1158 Forks: 0
Human Excitatory Cortical NGN2 Neuron Differentiation Protocol
*This protocol has been adapted from Zhang et al. 2013. It has two major parts: neural induction via overexpression of NGN2 and dual SMAD & Wnt inhibition and neuronal maturation. Plating density is to be titrated for each cell line as different lines have different viability.
1. Maintenance of ES/iPS cells:
Stem cells are grown on matrigel (Fisher Scientific #BD354277) coated TC dishes/plates and fed daily with mTeSR media (StemCell Technologies #05850). Cultures are passaged as required by accutase (Gibco, A11105) and replated in the presence of 10uM rock inhibitor (1:1000 of 1mM Stock) Y27632 (DNSK International, #129830-38-2) only for the first 24hrs.
2. Schematic
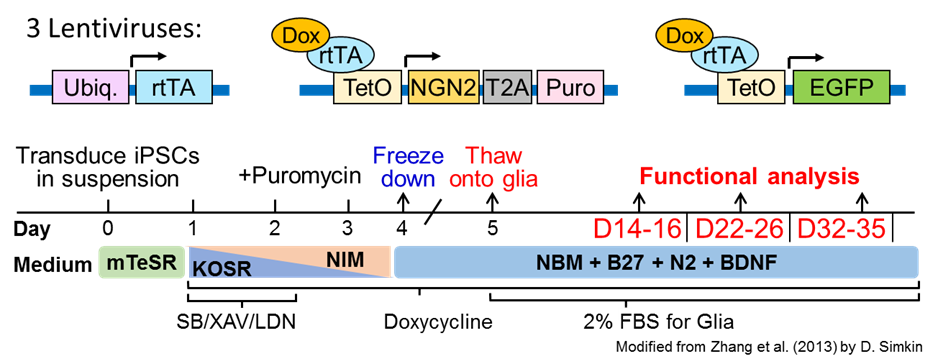
3. Differentiation protocol (example with 2 cell lines)
D0: - Prepare 2x 6-well plates (for differentiation) and 2x 10cm (to passage iPSCs normally) plates by coating all wells with marigel (30 min in hood then 30-60min in incubator). Prep the matrigel coated plates by washing with PBS and then adding 600 ul/well of 6-well or 10mL/ 10cm plate of mTeSR+Rock. Place plates in incubator till you are ready to plate.
- Mix the 3 lentiviruses together in one tube. For 12 wells add 2.4 mL (+ a drop or 2 for pipetting errors) of mTeSR + Rock (~200 µL/well) to a 15 mL conical tube and keep on ice. Add the 3 lentiviruses to this tube. Mix by inverting tube a few times and put back on ice for a few minutes and then split the lentivirus media into 2 separate tubes ~1245 µL each ((2.4 mL + lenti ~90 uL)/2). Let tubes sit on ice until you are ready to mix with cells.
| Lenti viruses (concentrated from NU lenti core): | Quantity per 1 million cells/1 well of 6-well (µL) | For 12 wells of 6-well or 12 million cells multiply by 12 (µL) |
FUW-M2rtTA Addgene #20342 | 4.1 µL | 49.2 µL |
| TetO-Ngn2-Puro Addgene #52047 | 2.9 µL | 34.8 µL |
TetO-EGFP Addgene #30130 | .6 µL | 7.2 µL |
- Passage iPSC (from almost confluent 10cm plate **) with Accutase and resuspend in (3-5 ml) mTeSR+ Rock inhibitor. Count cells (take average of 3 separate counts) and then transfer 0.95–1.1 million cells/well (depending on their rate of growth) to the virus tube. So for 6 wells per line transfer 6 to 6.6 million cells into the viral mixture tube and mix gently by pipetting up and down and then inverting a few times. You should have ~1245uL of virus plus whatever volume of cells you added in your tube which you will later split up to plate into 6 wells.
- Incubate cells/virus mixture in hood at room temp for 4-5 min (with lids slightly unscrewed for oxygen). Mix by inverting a few times half way through your 4-5 min wait (with lids closed!!). After closing the tube tap it a few times on the table to make sure liquid doesn’t get stuck in the cap.
- Calculate volume for 950K-1.1 million cells plus 200 µL (virus mixture). Then pipette cells/lenti mixture to wells. The aim is to have 60-70% confluence by the next day.
** Note: ALL new lenti batches must be titrated to know how much to use. The volumes indicated here are based on NU lenti core facility lenti prep titers.
** Note: (co-infection with TetO-GFP and rtTA but not TetO-Ngn2-Puro is typically used as a control). Virus amount varies with each prep and should be tested prior to scaling up. For a titer of 109, use 0.1ul of virus per 50,000 cells (always use more rtta though).* titer each virus batch before use!!!
** Note: In the case that stable iPSC lines have already been infected with the lenti viruses are used, just plate iPSCs without viral transduction and start D1 the following day.
** Note: 3 wells of a 6-well plate of stem cells will generate 5-8 million NGN2 neurons
D1: Feed (2 mls/well) with KOSR media + SB/XAV/LDN. Add Doxycyclin (1:6667; 1.5x) to induce NGN2 and GFP expression. Keep Dox in all media from this day onwards.
** Keep one well/line as stem cells and feed mTeSR until they are ready to passage to expand. Freeze ~1 million NGN2 transduced iPSCs and passage the rest onto 1 or 2 10cm plates. When these plates are almost confluent passage them again onto 2-4 10cm plates at 6 -7 million cells per plate (and make a freeze). The next day (D1) feed with D1 media (KOSR + SB/XAV/LDN plus Doxycyclin (1:6667; 1.5x)).
D2: Feed with 1:1 ratio of (KOSR + SB/XAV/LDN): (NIM) and add Dox (1:6667; 1.5x). Start selection by adding Puromycin (1:5000) to select for infected cells.
**Note: NIM is very sensitive and turns pink quickly which will affect differentiation qualityso make it from newly opened bottles of DMEM/F12, make small volumes andonly keep it for 2-3 days at 4⁰C.
D3: Feed with NIM media + Dox (1:6667; 1.5x), add Puromycin (1:5000).
D4: Freeze down: Use accutase 4-5min to dissociate cells, wash and spin down in neurobasal base media + puromycin (1:5000). Resuspend cells in NBM +Dox (1:6667; 1.5x), +BDNF and Rock inhibitor. Count cells (take average of 3 counts) and freeze down (in freezing container in -80C freezer) at 1-2 million cells per vial in 10% DMSO/Hyclone FBS. Move cells to liquid nitrogen within 2 days.
**Note: Freezes with higher density of neuron freezes tend to have higher after thaw viability but might be a waste of cells depending on experiment. Instead you can reduce the volume in which you freeze in (e.g. instead of 1ml freezes freeze in 500uL/vial).
For freezing neurons from 10cm plates make freezes of ~8-12 Million neurons per vial and whatever is left over so that you have relatively even numbers of high concentration vials. The goal is to have about 50-60 million neurons when you freeze down which equals about 30- 35 million at thaw. It's fine if you have less but it’s better to have multiple freezes of the same neurons for Q-state.
Viruses (viruses need to be titrated first since every batch may be at different concentration):
- TetO-Ngn2-Puro; addgene: #52047: use 2.8-80 uL/ 1 Million cells
- FUW-M2rtTA (Ubiq promotor); addgene #20342: use 3.8-80 uL/ 1 Million cells
- TetO-FUW-EGFP; addgene #30130: use .5-40 uL/ 1 Million cells
- Or TetO-FUW-RFP; Made by DSi: use .5-40 uL/ 1 Million cells
** Note: Only use high titer (>109) virus for infections
Thawing and plating NGN2 neurons
| Plate type (1 well) | # of Glia | # of Neurons | Purpose |
| 12mm coverslip in 24-well plate | 90K drop | 35-40K | ephys/imaging |
| 1w of 6-well | 200-250K | 450K-500K | qPCR/Western/sorting |
| 1w of 12-well MEA | 40-50K drop | 30-35K | MEA recording |
| 1w of 48-well MEA | 40-50K drop | 30-35K | MEA recording |
| 1w of 96-well Vala plate | 25-30K | 15K | imaging |
- Coat plates/coverslips with PDL overnight in 4°C. Next day wash plates x2 water and x1 with PBS and let dry completely (10-15 min in hood; make sure coverslip in the center of the well).
- Coat plates with 20 uM laminin diluted in PBS for at least 2 hour in incubator before plating mouse glia. When ready to plate glia aspirate laminin but DO NOT WASH IT, and let well dry a little bit while glia is spinning down (~2-4min).
- Plate mouse glia (P2) in glia media to make monolayer at least 4 days before neurons.
** Note: When plating glia, plate in volume of 50-70µl or 20µL drop of media, in center of coverslip or MEA well, respectively. Incubate 5-10 minutes for cells to adhere, then add 1 mL glia media on top.
D5: Thaw out frozen neuronsinto neurobasal +puro (1:5000)/ spin down and resuspend in NBM media + BDNF (1:10000), Dox (1:6667; 1.5x), 2% FBS and Rock inhibitor. Count and plate onto glia monolayer. Next day do half media change to NBM media + BDNF(1:10000), Dox (1:6667; 1.5x) + 2% FBS.
** Note: Usually you will get a little over half of the neurons frozen surviving the thaw.
D7-36: Continue to feed (half change) every 2-3 days (Mon, Wed, Fri) with NBM base media + BDNF/Dox (1:10000; 1x) + 2% FBS.
** Note: On Fridays make sure cells get extra media for the 2 day weekendand if neurons start to look clumpy (cells that didn’t get enough virus still dividing or glia disappearing) add laminin to media.
*** If there are too many progenitor or dividing cells taking over you can pulse Ara-C (1:10000) so Day 7 add Ara-C (1:5000 since doing half change) and then continue half-change feedings with normal media from then on (NBM + BDNF/Dox (1:10000; 1x) + 2% FBS).
Reagents: *It is important to stick to the same lot numbers for entire experiment because there may be differences between lot numbers of reagents.
| Reagent | Vendor | Cat no. | [Stock] | [Final] | Dilution |
| LDN-193189 | DNSK International | 1062368-24-4 | 1 mM | 100 nM | 1:10000 |
| SB431542 | Tocris | 1254 | 10 mM | 10 uM | 1:1000 |
| XAV939 | Stemgent | 04-00046 | 10 mM | 2 uM | 1:5000 |
| Doxycycline | Sigma Aldrich | D9891-1G | 20 mg/mL | 2 ug/mL | 1:10000 |
| Puromycin | Sigma-Aldrich | P8833 | 10 mg/mL | 2 ug/mL | 1:5000 |
| BDNF | R&D Systems | 248-BD | 100 ug/mL | 10 ng/mL | 1:10000 |
| Laminin | Life Technologies | 23017-015 | 1 mg/mL | 20 ug/mL | 1:50 |
| Heparin Sulfate | Sigma Aldrich | H3149 | 50 mg/mL | 2 ug/mL | 1:25000 |
| BME (diluted) | Gibco™ (thermofisher) | 21985023 | 55 mM | 55ul | 1:1000 |
| ***Ara-C | Sigma Aldrich | C1768 | 40mM | 4uM | 1:10000 |
Media Recipes:
KOSR (Knockout Serum Replacement Medium) scaled down to 50 mL
41.5 mL Knockout DMEM (Life Technologies # 10829-018)
7.5 mL Knockout Replacement Serum KSR (15%) (Life Technologies # 10828028)
500 µL MEM non-essential amino acids (NEAA; Life Technologies # 10370088)
500 µL Glutamax (Life Technologies # 35050061)
50 µLbeta-mercaptoethanol (diluted stock)
Add BME after sterile filtering, store for <2 weeks 4°C, light protected
NIM (Neural Induction Medium) scaled down to 50 mL (NIM tends to turn pink very fast)
48.1 mL DMEM/F12 + L-glutamine (Life Technologies #11320-033)
500 µL Glutamax
500 µL NEAA
400 µL 20% D-glucose in H2O (Sigma Aldrich # G8769-100ML 45% D-glucose)
500 µL N2 supplement (Life Technologies # 17502048)
2 µL Heparin Sulfate (HS; 2 ug/ml final conc.)
Add N2 and Heparin Sulfate after sterile filtering, store for <1 week 4°C, light protected
NBM (NeuroBasal Medium): 500 mL
475 mL Neurobasal+ L-glutamine (Life Technologies # 21103049)
5 mL Glutamax
5 mL NEAA
5 mL N2 supplement
10 mL B27 supplement (Life Technologies # 17504044)
Add N2 and B27 after sterile filtering, store for <2 weeks 4°C, light protected
Glia Media: 500 mL
445 mL DMEM (cat #: 15013CV)
5 mL Glutamax (cat #: 35050061)
50 mL HI Horse Serum (cat#: 26050-088)
Filter store at 4°C for <3 weeks or if it turns pink. Change glia media at least x1 a week.
- Simkin, D(2021). Cortical excitatory neuron differentiation. Bio-protocol Preprint. bio-protocol.org/prep981.
- Simkin, D., Marshall, K. A., Vanoye, C. G., Desai, R. R., Bustos, B. I., Piyevsky, B. N., Ortega, J. A., Forrest, M., Robertson, G. L., Penzes, P., Laux, L. C., Lubbe, S. J., Millichap, J. J., George, A. L. and Kiskinis, E.(2021). Dyshomeostatic modulation of Ca2+-activated K+ channels in a human neuronal model of KCNQ2 encephalopathy. eLife. DOI: 10.7554/eLife.64434
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link
